Sertoli cell survival and barrier function are regulated by miR-181c/d-Pafah1b1 axis during mammalian spermatogenesis
- PMID: 36008729
- PMCID: PMC9411099
- DOI: 10.1007/s00018-022-04521-w
Sertoli cell survival and barrier function are regulated by miR-181c/d-Pafah1b1 axis during mammalian spermatogenesis
Abstract
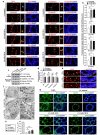
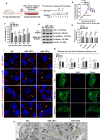
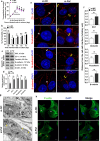
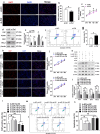
Sertoli cells contribute to the formation of the blood-testis barrier (BTB), which is necessary for normal spermatogenesis. Recently, microRNAs (miRNAs) have emerged as posttranscriptional regulatory elements in BTB function during spermatogenesis. Our previous study has shown that miR-181c or miR-181d (miR-181c/d) is highly expressed in testes from boars at 60 days old compared with at 180 days old. Herein, we found that overexpression of miR-181c/d via miR-181c/d mimics in murine Sertoli cells (SCs) or through injecting miR-181c/d-overexpressing lentivirus in murine testes perturbs BTB function by altering BTB-associated protein distribution at the Sertoli cell-cell interface and F-actin organization, but this in vivo perturbation disappears approximately 6 weeks after the final treatment. We also found that miR-181c/d represses Sertoli cell proliferation and promotes its apoptosis. Moreover, miR-181c/d regulates Sertoli cell survival and barrier function by targeting platelet-activating factor acetylhydrolase 1b regulatory subunit 1 (Pafah1b1) gene. Furthermore, miR-181c/d suppresses PAFAH1B1 expression, reduces the complex of PAFAH1B1 with IQ motif-containing GTPase activating protein 1, and inhibits CDC42/PAK1/LIMK1/Cofilin pathway which is required for F-actin stabilization. In total, our results reveal the regulatory axis of miR-181c/d-Pafah1b1 in cell survival and barrier function of Sertoli cells and provide additional insights into miRNA functions in mammalian spermatogenesis.
Keywords: Blood-testis barrier; Ectoplasmic specialization; Mammals; Pafah1b1; Sertoli cells; Spermatogenesis; Tight junction; miR-181c/d.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures


Similar articles
-
rpS6 Regulates blood-testis barrier dynamics by affecting F-actin organization and protein recruitment.Endocrinology. 2012 Oct;153(10):5036-48. doi: 10.1210/en.2012-1665. Epub 2012 Sep 4. Endocrinology. 2012. PMID: 22948214 Free PMC article.
-
MARK2 and MARK4 Regulate Sertoli Cell BTB Dynamics Through Microtubule and Actin Cytoskeletons.Endocrinology. 2022 Oct 11;163(11):bqac130. doi: 10.1210/endocr/bqac130. Endocrinology. 2022. PMID: 35971301 Free PMC article.
-
Regulation of spermatogenesis by a local functional axis in the testis: role of the basement membrane-derived noncollagenous 1 domain peptide.FASEB J. 2017 Aug;31(8):3587-3607. doi: 10.1096/fj.201700052R. Epub 2017 May 9. FASEB J. 2017. PMID: 28487282 Free PMC article.
-
Regulation of Blood-Testis Barrier (BTB) Dynamics, Role of Actin-, and Microtubule-Based Cytoskeletons.Methods Mol Biol. 2018;1748:229-243. doi: 10.1007/978-1-4939-7698-0_16. Methods Mol Biol. 2018. PMID: 29453575 Free PMC article. Review.
-
Is toxicant-induced Sertoli cell injury in vitro a useful model to study molecular mechanisms in spermatogenesis?Semin Cell Dev Biol. 2016 Nov;59:141-156. doi: 10.1016/j.semcdb.2016.01.003. Epub 2016 Jan 15. Semin Cell Dev Biol. 2016. PMID: 26779951 Free PMC article. Review.
Cited by
-
DCAF2 regulates the proliferation and differentiation of mouse progenitor spermatogonia by targeting p21 and thymine DNA glycosylase.Cell Prolif. 2024 Oct;57(10):e13676. doi: 10.1111/cpr.13676. Epub 2024 Jun 4. Cell Prolif. 2024. PMID: 38837535 Free PMC article.
-
Inhibition of miR-143-3p Restores Blood-Testis Barrier Function and Ameliorates Sertoli Cell Senescence.Cells. 2024 Feb 8;13(4):313. doi: 10.3390/cells13040313. Cells. 2024. PMID: 38391926 Free PMC article.
-
[Research progress on glycolipid metabolism of Sertoli cell in the development of spermatogenic cell].Zhejiang Da Xue Xue Bao Yi Xue Ban. 2025 Mar 25;54(2):257-265. doi: 10.3724/zdxbyxb-2024-0346. Zhejiang Da Xue Xue Bao Yi Xue Ban. 2025. PMID: 40065698 Free PMC article. Review. Chinese.
-
The Role of microRNA in Spermatogenesis: Is There a Place for Fertility Preservation Innovation?Int J Mol Sci. 2023 Dec 29;25(1):460. doi: 10.3390/ijms25010460. Int J Mol Sci. 2023. PMID: 38203631 Free PMC article. Review.
-
The microRNA-mediated apoptotic signaling axis in male reproduction: a possible and targetable culprit in male infertility.Cell Biol Toxicol. 2025 Mar 5;41(1):54. doi: 10.1007/s10565-025-10006-w. Cell Biol Toxicol. 2025. PMID: 40038116 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

