Capturing the antibiotic resistome of preterm infants reveals new benefits of probiotic supplementation
- PMID: 36008821
- PMCID: PMC9414150
- DOI: 10.1186/s40168-022-01327-7
Capturing the antibiotic resistome of preterm infants reveals new benefits of probiotic supplementation
Abstract
Background: Probiotic use in preterm infants can mitigate the impact of antibiotic exposure and reduce rates of certain illnesses; however, the benefit on the gut resistome, the collection of antibiotic resistance genes, requires further investigation. We hypothesized that probiotic supplementation of early preterm infants (born < 32-week gestation) while in hospital reduces the prevalence of antibiotic resistance genes associated with pathogenic bacteria in the gut. We used a targeted capture approach to compare the resistome from stool samples collected at the term corrected age of 40 weeks for two groups of preterm infants (those that routinely received a multi-strain probiotic during hospitalization and those that did not) with samples from full-term infants at 10 days of age to identify if preterm birth or probiotic supplementation impacted the resistome. We also compared the two groups of preterm infants up to 5 months of age to identify persistent antibiotic resistance genes.
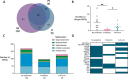
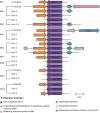
Results: At the term corrected age, or 10 days of age for the full-term infants, we found over 80 antibiotic resistance genes in the preterm infants that did not receive probiotics that were not identified in either the full-term or probiotic-supplemented preterm infants. More genes associated with antibiotic inactivation mechanisms were identified in preterm infants unexposed to probiotics at this collection time-point compared to the other infants. We further linked these genes to mobile genetic elements and Enterobacteriaceae, which were also abundant in their gut microbiomes. Various genes associated with aminoglycoside and beta-lactam resistance, commonly found in pathogenic bacteria, were retained for up to 5 months in the preterm infants that did not receive probiotics.
Conclusions: This pilot survey of preterm infants shows that probiotics administered after preterm birth during hospitalization reduced the diversity and prevented persistence of antibiotic resistance genes in the gut microbiome. The benefits of probiotic use on the microbiome and the resistome should be further explored in larger groups of infants. Due to its high sensitivity and lower sequencing cost, our targeted capture approach can facilitate these surveys to further address the implications of resistance genes persisting into infancy without the need for large-scale metagenomic sequencing. Video Abstract.
Keywords: Antibiotics; Gut microbiota; Preterm infants; Probiotics; Resistome; Targeted capture.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures




References
-
- van den Akker CHP, van Goudoever JB, Shamir R, Domellöf M, Embleton ND, Hojsak I, et al. Probiotics and preterm infants: a position paper by the European Society for Paediatric Gastroenterology Hepatology and Nutrition Committee on Nutrition and the European Society for Paediatric Gastroenterology Hepatology and Nutrition Working Group for Pr. J Pediatr Gastroenterol Nutr. 2020;70:664–680. doi: 10.1097/MPG.0000000000002655. - DOI - PubMed
-
- Simioni J, Hutton EK, Gunn E, Holloway AC, Stearns JC, McDonald H, et al. A comparison of intestinal microbiota in a population of low-risk infants exposed and not exposed to intrapartum antibiotics: the Baby & Microbiota of the Intestine Cohort Study protocol. BMC Pediatr. 2016;16:1–7. doi: 10.1186/s12887-016-0724-5. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

