A Trajectory of Discovery: Metabolic Regulation by the Conditionally Disordered Chloroplast Protein, CP12
- PMID: 36008940
- PMCID: PMC9406205
- DOI: 10.3390/biom12081047
A Trajectory of Discovery: Metabolic Regulation by the Conditionally Disordered Chloroplast Protein, CP12
Abstract
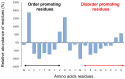
The chloroplast protein CP12, which is widespread in photosynthetic organisms, belongs to the intrinsically disordered proteins family. This small protein (80 amino acid residues long) presents a bias in its composition; it is enriched in charged amino acids, has a small number of hydrophobic residues, and has a high proportion of disorder-promoting residues. More precisely, CP12 is a conditionally disordered proteins (CDP) dependent upon the redox state of its four cysteine residues. During the day, reducing conditions prevail in the chloroplast, and CP12 is fully disordered. Under oxidizing conditions (night), its cysteine residues form two disulfide bridges that confer some stability to some structural elements. Like many CDPs, CP12 plays key roles, and its redox-dependent conditional disorder is important for the main function of CP12: the dark/light regulation of the Calvin-Benson-Bassham (CBB) cycle responsible for CO2 assimilation. Oxidized CP12 binds to glyceraldehyde-3-phosphate dehydrogenase and phosphoribulokinase and thereby inhibits their activity. However, recent studies reveal that CP12 may have other functions beyond the CBB cycle regulation. In this review, we report the discovery of this protein, its features as a disordered protein, and the many functions this small protein can have.
Keywords: Calvin-Benson-Bassham cycle; conditionally disordered protein; history of modern science; metabolism regulation; moonlighting protein; protein-protein interaction.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

