Targeting the YXXΦ Motifs of the SARS Coronaviruses 1 and 2 ORF3a Peptides by In Silico Analysis to Predict Novel Virus-Host Interactions
- PMID: 36008946
- PMCID: PMC9405953
- DOI: 10.3390/biom12081052
Targeting the YXXΦ Motifs of the SARS Coronaviruses 1 and 2 ORF3a Peptides by In Silico Analysis to Predict Novel Virus-Host Interactions
Abstract
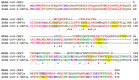
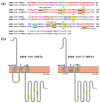
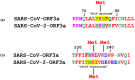
The emerging SARS-CoV and SARS-CoV-2 belong to the family of "common cold" RNA coronaviruses, and they are responsible for the 2003 epidemic and the current pandemic with over 6.3 M deaths worldwide. The ORF3a gene is conserved in both viruses and codes for the accessory protein ORF3a, with unclear functions, possibly related to viral virulence and pathogenesis. The tyrosine-based YXXΦ motif (Φ: bulky hydrophobic residue-L/I/M/V/F) was originally discovered to mediate clathrin-dependent endocytosis of membrane-spanning proteins. Many viruses employ the YXXΦ motif to achieve efficient receptor-guided internalisation in host cells, maintain the structural integrity of their capsids and enhance viral replication. Importantly, this motif has been recently identified on the ORF3a proteins of SARS-CoV and SARS-CoV-2. Given that the ORF3a aa sequence is not fully conserved between the two SARS viruses, we aimed to map in silico structural differences and putative sequence-driven alterations of regulatory elements within and adjacently to the YXXΦ motifs that could predict variations in ORF3a functions. Using robust bioinformatics tools, we investigated the presence of relevant post-translational modifications and the YXXΦ motif involvement in protein-protein interactions. Our study suggests that the predicted YXXΦ-related features may confer specific-yet to be discovered-functions to ORF3a proteins, significant to the new virus and related to enhanced propagation, host immune regulation and virulence.
Keywords: ORF3a; SARS-CoV; SARS-CoV-2; YXXΦ motif; immune response; post-translational modifications.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

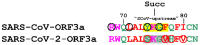

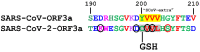

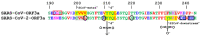
Similar articles
-
Comprehensive computational analysis reveals YXXΦ[I/L/M/F/V] motif and YXXΦ-like tetrapeptides across HFRS causing Hantaviruses and their association with viral pathogenesis and host immune regulation.Front Immunol. 2022 Oct 6;13:1031608. doi: 10.3389/fimmu.2022.1031608. eCollection 2022. Front Immunol. 2022. PMID: 36275660 Free PMC article.
-
A novel diG motif in ORF3a protein of SARS-Cov-2 for intracellular transport.Front Cell Dev Biol. 2022 Nov 23;10:1011221. doi: 10.3389/fcell.2022.1011221. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 36506095 Free PMC article.
-
Genome-Wide Characterization of SARS-CoV-2 Cytopathogenic Proteins in the Search of Antiviral Targets.mBio. 2021 Feb 22;13(1):e0016922. doi: 10.1128/mbio.00169-22. Epub 2022 Feb 15. mBio. 2021. PMID: 35164548 Free PMC article.
-
Understanding the Role of SARS-CoV-2 ORF3a in Viral Pathogenesis and COVID-19.Front Microbiol. 2022 Mar 9;13:854567. doi: 10.3389/fmicb.2022.854567. eCollection 2022. Front Microbiol. 2022. PMID: 35356515 Free PMC article. Review.
-
Current status of antivirals and druggable targets of SARS CoV-2 and other human pathogenic coronaviruses.Drug Resist Updat. 2020 Dec;53:100721. doi: 10.1016/j.drup.2020.100721. Epub 2020 Aug 26. Drug Resist Updat. 2020. PMID: 33132205 Free PMC article. Review.
Cited by
-
Comprehensive computational analysis reveals YXXΦ[I/L/M/F/V] motif and YXXΦ-like tetrapeptides across HFRS causing Hantaviruses and their association with viral pathogenesis and host immune regulation.Front Immunol. 2022 Oct 6;13:1031608. doi: 10.3389/fimmu.2022.1031608. eCollection 2022. Front Immunol. 2022. PMID: 36275660 Free PMC article.
-
Endoplasmic reticulum-associated SARS-CoV-2 ORF3a elicits heightened cytopathic effects despite robust ER-associated degradation.mBio. 2024 Jan 16;15(1):e0303023. doi: 10.1128/mbio.03030-23. Epub 2023 Dec 11. mBio. 2024. PMID: 38078754 Free PMC article.
-
SARS-CoV-2 ORF3a Protein as a Therapeutic Target against COVID-19 and Long-Term Post-Infection Effects.Pathogens. 2024 Jan 14;13(1):75. doi: 10.3390/pathogens13010075. Pathogens. 2024. PMID: 38251382 Free PMC article. Review.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

