GraphSite: Ligand Binding Site Classification with Deep Graph Learning
- PMID: 36008947
- PMCID: PMC9405584
- DOI: 10.3390/biom12081053
GraphSite: Ligand Binding Site Classification with Deep Graph Learning
Abstract
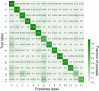
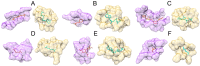
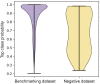
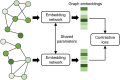
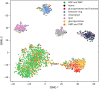
The binding of small organic molecules to protein targets is fundamental to a wide array of cellular functions. It is also routinely exploited to develop new therapeutic strategies against a variety of diseases. On that account, the ability to effectively detect and classify ligand binding sites in proteins is of paramount importance to modern structure-based drug discovery. These complex and non-trivial tasks require sophisticated algorithms from the field of artificial intelligence to achieve a high prediction accuracy. In this communication, we describe GraphSite, a deep learning-based method utilizing a graph representation of local protein structures and a state-of-the-art graph neural network to classify ligand binding sites. Using neural weighted message passing layers to effectively capture the structural, physicochemical, and evolutionary characteristics of binding pockets mitigates model overfitting and improves the classification accuracy. Indeed, comprehensive cross-validation benchmarks against a large dataset of binding pockets belonging to 14 diverse functional classes demonstrate that GraphSite yields the class-weighted F1-score of 81.7%, outperforming other approaches such as molecular docking and binding site matching. Further, it also generalizes well to unseen data with the F1-score of 70.7%, which is the expected performance in real-world applications. We also discuss new directions to improve and extend GraphSite in the future.
Keywords: deep learning; graph neural network; ligand binding sites; structure-based drug discovery.
Conflict of interest statement
The authors declare no conflict of interests.
Figures


References
-
- Armstrong J.D., Hubbard R.E., Farrell T., Maiguashca B., editors. Structure-Based Drug Discovery: An Overview. The Royal Society of Chemistry; Cambridge, UK: 2006.
-
- Vos T., Lim S.S., Abbafati C., Abbas K.M., Abbasi M., Abbasifard M., Abbasi-Kangevari M., Abbastabar H., Abd-Allah F., Abdelalim A., et al. Global burden of 369 diseases and injuries in 204 countries and territories, 1990–2019: A systematic analysis for the Global Burden of Disease Study 2019. Lancet. 2020;396:1204–1222. doi: 10.1016/S0140-6736(20)30925-9. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

