Isolation, Characterization, and Autophagy Function of BECN1-Splicing Isoforms in Cancer Cells
- PMID: 36008963
- PMCID: PMC9405542
- DOI: 10.3390/biom12081069
Isolation, Characterization, and Autophagy Function of BECN1-Splicing Isoforms in Cancer Cells
Abstract
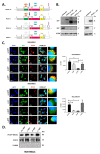
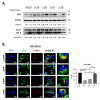
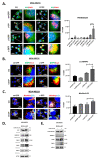
Alternative splicing allows the synthesis of different protein variants starting from a single gene. Human Beclin 1 (BECN1) is a key autophagy regulator that acts as haploinsufficient tumor suppressor since its decreased expression correlates with tumorigenesis and poor prognosis in cancer patients. Recent studies show that BECN1 mRNA undergoes alternative splicing. Here, we report on the isolation and molecular and functional characterization of three BECN1 transcript variants (named BECN1-α, -β and -γ) in human cancer cells. In ovarian cancer NIHOVCAR3, these splicing variants were found along with the canonical wild-type. BECN1-α lacks 143 nucleotides at its C-terminus and corresponds to a variant previously described. BECN1-β and -γ lack the BCL2 homology 3 domain and other regions at their C-termini. Following overexpression in breast cancer cells MDA-MB231, we found that BECN1-α stimulates autophagy. Specifically, BECN1-α binds to Parkin and stimulates mitophagy. On the contrary, BECN1-β reduces autophagy with a dominant negative effect over the endogenous wild-type isoform. BECN1-γ maintains its ability to interact with the vacuolar protein sorting 34 and only has a slight effect on autophagy. It is possible that cancer cells utilize the alternative splicing of BECN1 for modulating autophagy and mitophagy in response to environmental stresses.
Keywords: BH3 domain breast cancers; Bcl-2; Beclin 1; alternative splicing; autophagy; isoforms; mitophagy; ovarian cancers.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

