Low Complexity Induces Structure in Protein Regions Predicted as Intrinsically Disordered
- PMID: 36008992
- PMCID: PMC9405754
- DOI: 10.3390/biom12081098
Low Complexity Induces Structure in Protein Regions Predicted as Intrinsically Disordered
Abstract
There is increasing evidence that many intrinsically disordered regions (IDRs) in proteins play key functional roles through interactions with other proteins or nucleic acids. These interactions often exhibit a context-dependent structural behavior. We hypothesize that low complexity regions (LCRs), often found within IDRs, could have a role in inducing local structure in IDRs. To test this, we predicted IDRs in the human proteome and analyzed their structures or those of homologous sequences in the Protein Data Bank (PDB). We then identified two types of simple LCRs within IDRs: regions with only one (polyX or homorepeats) or with only two types of amino acids (polyXY). We were able to assign structural information from the PDB more often to these LCRs than to the surrounding IDRs (polyX 61.8% > polyXY 50.5% > IDRs 39.7%). The most frequently observed polyX and polyXY within IDRs contained E (Glu) or G (Gly). Structural analyses of these sequences and of homologs indicate that polyEK regions induce helical conformations, while the other most frequent LCRs induce coil structures. Our work proposes bioinformatics methods to help in the study of the structural behavior of IDRs and provides a solid basis suggesting a structuring role of LCRs within them.
Keywords: homorepeats; intrinsically disordered regions; low complexity regions; protein structure.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures
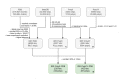
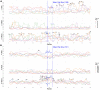

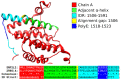



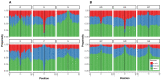
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

