Mitochondria Targeted Antioxidant Significantly Alleviates Preeclampsia Caused by 11β-HSD2 Dysfunction via OPA1 and MtDNA Maintenance
- PMID: 36009224
- PMCID: PMC9404992
- DOI: 10.3390/antiox11081505
Mitochondria Targeted Antioxidant Significantly Alleviates Preeclampsia Caused by 11β-HSD2 Dysfunction via OPA1 and MtDNA Maintenance
Abstract
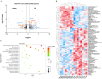
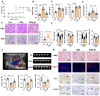
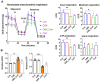
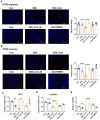
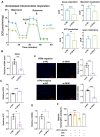
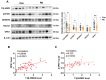
We have previously demonstrated that placental 11β-hydroxysteroid dehydrogenase type 2 (11β-HSD2) dysfunction contributes to PE pathogenesis. We sought to elucidate molecular mechanisms underlying 11β-HSD2 dysfunction-induced PE and to seek potential therapeutic targets using a 11β-HSD2 dysfunction-induced PE-like rat model as well as cultured extravillous trophoblasts (EVTs) since PE begins with impaired function of EVTs. In 11β-HSD2 dysfunction-induced PE-like rat model, we revealed that placental mitochondrial dysfunction occurred, which was associated with mitDNA instability and impaired mitochondrial dynamics, such as decreased optic atrophy 1 (OPA1) expression. MitoTEMPO treatment significantly alleviated the hallmark of PE-like features and improved mitDNA stability and mitochondrial dynamics in the placentas of rat PE-like model. In cultured human EVTs, we found that 11β-HSD2 dysfunction led to mitochondrial dysfunction and disrupted mtDNA stability. MitoTEMPO treatment improved impaired invasion and migration induced by 11β-HSD2 dysfunction in cultured EVTs. Further, we revealed that OPA1 was one of the key factors that mediated 11β-HSD2 dysfunction-induced excess ROS production, mitochondrial dysfunction and mtDNA reduction. Our data indicates that 11β-HSD2 dysfunction causes mitochondrial dysfunctions, which impairs trophoblast function and subsequently results in PE development. Our study immediately highlights that excess ROS is a potential therapeutic target for PE.
Keywords: OXPHOS; mitochondria; mtDNA; placenta; preeclampsia.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Contribution of placental 11β-HSD2 to the pathogenesis of preeclampsia.FASEB J. 2020 Nov;34(11):15379-15399. doi: 10.1096/fj.202001003RR. Epub 2020 Sep 25. FASEB J. 2020. PMID: 32978833
-
Reduced expression of 11β-hydroxysteroid dehydrogenase type 2 in preeclamptic placentas is associated with decreased PPARγ but increased PPARα expression.Endocrinology. 2014 Jan;155(1):299-309. doi: 10.1210/en.2013-1350. Epub 2013 Dec 4. Endocrinology. 2014. PMID: 24169559
-
The ERK1/2 signaling pathway regulates 11beta-hydroxysteroid dehydrogenase type 2 expression in human trophoblast cells through a transcriptional mechanism.Biol Reprod. 2013 Oct 17;89(4):92. doi: 10.1095/biolreprod.113.110924. Print 2013 Oct. Biol Reprod. 2013. PMID: 23966319
-
Foetal and placental 11β-HSD2: a hub for developmental programming.Acta Physiol (Oxf). 2014 Feb;210(2):288-95. doi: 10.1111/apha.12187. Epub 2013 Dec 12. Acta Physiol (Oxf). 2014. PMID: 24330050 Review.
-
11beta-Hydroxysteroid dehydrogenase type 2 in pregnancy and preeclampsia.Mol Aspects Med. 2007 Apr;28(2):220-6. doi: 10.1016/j.mam.2007.04.003. Epub 2007 Apr 27. Mol Aspects Med. 2007. PMID: 17532462 Review.
Cited by
-
Pregnancy Metabolic Adaptation and Changes in Placental Metabolism in Preeclampsia.Geburtshilfe Frauenheilkd. 2024 Sep 19;84(11):1033-1042. doi: 10.1055/a-2403-4855. eCollection 2024 Nov. Geburtshilfe Frauenheilkd. 2024. PMID: 39524034 Free PMC article.
-
[Research progress of optic atrophy 1-mediated mitochondrial dynamics in skeletal system diseases].Zhongguo Xiu Fu Chong Jian Wai Ke Za Zhi. 2023 Jun 15;37(6):758-763. doi: 10.7507/1002-1892.202302056. Zhongguo Xiu Fu Chong Jian Wai Ke Za Zhi. 2023. PMID: 37331956 Free PMC article. Review. Chinese.
-
Estrogen receptors in mitochondrial metabolism: age-related changes and implications for pregnancy complications.Aging Adv. 2024 Dec;1(2):154-171. doi: 10.4103/agingadv.agingadv-d-24-00012. Epub 2024 Dec 20. Aging Adv. 2024. PMID: 39839811 Free PMC article.
-
Multiple analytical perspectives of mitochondrial genes in the context of preeclampsia: potential diagnostic markers.Front Immunol. 2025 Jul 17;16:1595706. doi: 10.3389/fimmu.2025.1595706. eCollection 2025. Front Immunol. 2025. PMID: 40746540 Free PMC article.
-
Mitochondrial ROS Accumulation Contributes to Maternal Hypertension and Impaired Remodeling of Spiral Artery but Not IUGR in a Rat PE Model Caused by Maternal Glucocorticoid Exposure.Antioxidants (Basel). 2023 Apr 24;12(5):987. doi: 10.3390/antiox12050987. Antioxidants (Basel). 2023. PMID: 37237853 Free PMC article.
References
LinkOut - more resources
Full Text Sources

