miRNAs and isomiRs: Serum-Based Biomarkers for the Development of Intellectual Disability and Autism Spectrum Disorder in Tuberous Sclerosis Complex
- PMID: 36009385
- PMCID: PMC9405248
- DOI: 10.3390/biomedicines10081838
miRNAs and isomiRs: Serum-Based Biomarkers for the Development of Intellectual Disability and Autism Spectrum Disorder in Tuberous Sclerosis Complex
Abstract
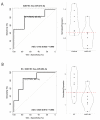
Tuberous sclerosis complex (TSC) is a rare multi-system genetic disorder characterized by a high incidence of epilepsy and neuropsychiatric manifestations known as tuberous-sclerosis-associated neuropsychiatric disorders (TANDs), including autism spectrum disorder (ASD) and intellectual disability (ID). MicroRNAs (miRNAs) are small regulatory non-coding RNAs that regulate the expression of more than 60% of all protein-coding genes in humans and have been reported to be dysregulated in several diseases, including TSC. In the current study, RNA sequencing analysis was performed to define the miRNA and isoform (isomiR) expression patterns in serum. A Receiver Operating Characteristic (ROC) curve analysis was used to identify circulating molecular biomarkers, miRNAs, and isomiRs, able to discriminate the development of neuropsychiatric comorbidity, either ASD, ID, or ASD + ID, in patients with TSC. Part of our bioinformatics predictions was verified with RT-qPCR performed on RNA isolated from patients' serum. Our results support the notion that circulating miRNAs and isomiRs have the potential to aid standard clinical testing in the early risk assessment of ASD and ID development in TSC patients.
Keywords: autism spectrum disorder; biomarkers; epilepsy; intellectual disability; serum; tuberous sclerosis complex.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Northrup H., Aronow M.E., Bebin E.M., Bissler J., Darling T.N., de Vries P.J., Frost M.D., Fuchs Z., Gosnell E.S., Gupta N., et al. Updated International Tuberous Sclerosis Complex Diagnostic Criteria and Surveillance and Management Recommendations. Pediatr. Neurol. 2021;123:50–66. doi: 10.1016/j.pediatrneurol.2021.07.011. - DOI - PubMed
-
- Sancak O., Nellist M., Goedbloed M., Elfferich P., Wouters C., Maat-Kievit A., Zonnenberg B., Verhoef S., Halley D., van den Ouweland A. Mutational Analysis of the TSC1 and TSC2 Genes in a Diagnostic Setting: Genotype--Phenotype Correlations and Comparison of Diagnostic DNA Techniques in Tuberous Sclerosis Complex. Eur. J. Hum. Genet. EJHG. 2005;13:731–741. doi: 10.1038/sj.ejhg.5201402. - DOI - PubMed
-
- Capal J.K., Williams M.E., Pearson D.A., Kissinger R., Horn P.S., Murray D., Currans K., Kent B., Bebin M., Northrup H., et al. Profile of Autism Spectrum Disorder in Tuberous Sclerosis Complex: Results from a Longitudinal, Prospective, Multisite Study. Ann. Neurol. 2021;90:874–886. doi: 10.1002/ana.26249. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

