The Journey of Human Transthyretin: Synthesis, Structure Stability, and Catabolism
- PMID: 36009453
- PMCID: PMC9405911
- DOI: 10.3390/biomedicines10081906
The Journey of Human Transthyretin: Synthesis, Structure Stability, and Catabolism
Abstract
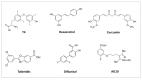
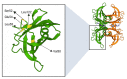
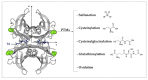
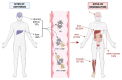
Transthyretin (TTR) is a homotetrameric protein mainly synthesised by the liver and the choroid plexus whose function is to carry the thyroid hormone thyroxine and the retinol-binding protein bound to retinol in plasma and cerebrospinal fluid. When the stability of the tetrameric structure is lost, it breaks down, paving the way for the aggregation of TTR monomers into insoluble fibrils leading to transthyretin (ATTR) amyloidosis, a progressive disorder mainly affecting the heart and nervous system. Several TTR gene mutations have been characterised as destabilisers of TTR structure and are associated with hereditary forms of ATTR amyloidosis. The reason why also the wild-type TTR is intrinsically amyloidogenic in some subjects is largely unknown. The aim of the review is to give an overview of the TTR biological life cycle which is largely unknown. For this purpose, the current knowledge on TTR physiological metabolism, from its synthesis to its catabolism, is described. Furthermore, a large section of the review is dedicated to examining in depth the role of mutations and physiological ligands on the stability of TTR tetramers.
Keywords: ER-associated degradation pathway; TTR amyloidosis; TTR clearance; retinol; retinol-binding protein; thyroxine; transthyretin.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Nilsson S., Rask L., Peterson P. Studies on thyroid hormone-binding proteins. II. Binding of thyroid hormones, retinol-binding protein, and fluorescent probes to prealbumin and effects of thyroxine on prealbumin subunit self association. J. Biol. Chem. 1975;250:8554–8563. doi: 10.1016/S0021-9258(19)40795-3. - DOI - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

