Genomic Aberrations Generate Fusion Gene FOXK2::TP63 and Activate NFKB1 in Cutaneous T-Cell Lymphoma
- PMID: 36009586
- PMCID: PMC9406051
- DOI: 10.3390/biomedicines10082038
Genomic Aberrations Generate Fusion Gene FOXK2::TP63 and Activate NFKB1 in Cutaneous T-Cell Lymphoma
Abstract
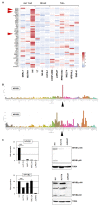
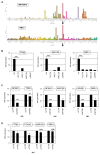
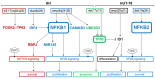
Cutaneous T-cell lymphoma (CTCL) is a severe lymphoid malignancy with a worse prognosis lacking curative treatment regimens. Several gene mutations and deregulated pathways, including NFkB signaling, have been implicated in its pathogenesis. Accordingly, CTCL cell line HUT-78 reportedly contains mutated NFKB2, which is constitutively activated via partial gene deletion, also demonstrating that genomic rearrangements cause driving mutations in this malignancy. Here, along with HUT-78, we analyzed CTCL cell line HH to identify additional aberrations underlying gene deregulation. Karyotyping and genomic profiling of HH showed several rearrangements worthy of detailed investigation. Corresponding to the established karyotype, RNA-seq data and PCR analysis confirmed the presence of t(3;17)(q28;q25), generating a novel fusion gene, FOXK2::TP63. Furthermore, chromosomal rearrangement t(1;4)(p32;q25) was connected to amplification at 4q24-26, affecting aberrant NFKB1 overexpression thereat. Transcription factor binding-site analysis and knockdown experiments demonstrated that IRF4 contributed to NFKB1 expression. Within the same amplicon, we identified amplification and overexpression of NFkB signaling activator CAMK2D (4q26) and p53-inhibitor UBE2D3 (4q24). Genomic profiling data for HUT-78 detailed a deletion at 10q25 underlying reported NFKB2 activation. Moreover, amplifications of ID1 (20q11) and IKZF2 (2q34) in this cell line drove overexpression of these NK cell differentiation factors and possibly thus formed corresponding lineage characteristics. Target gene analysis for NFKB1 via siRNA-mediated knockdown in HH revealed activation of TP63, MIR155, and NOTCH pathway component RBPJ. Finally, treatment of HH with NFkB inhibitor demonstrated a role for NFkB in supporting proliferation, while usage of inhibitor DAPT showed significant survival effects via the NOTCH pathway. Collectively, our data suggest that NFkB and/or NOTCH inhibitors may represent reasonable treatment options for subsets of CTCL patients.
Keywords: MOTN-1; T-ALL; TPL1XR1::TP63.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






Similar articles
-
Heterogeneous chromosomal aberrations generate 3' truncations of the NFKB2/lyt-10 gene in lymphoid malignancies.Blood. 1994 Dec 1;84(11):3850-60. Blood. 1994. PMID: 7949142
-
PAK1 overexpression promotes cell proliferation in cutaneous T cell lymphoma via suppression of PUMA and p21.J Dermatol Sci. 2018 Apr;90(1):60-67. doi: 10.1016/j.jdermsci.2017.11.019. Epub 2018 Jan 4. J Dermatol Sci. 2018. PMID: 29307600
-
Rearrangement and altered expression of the NFKB-2 gene in human cutaneous T-lymphoma cells.Oncogene. 1994 Aug;9(8):2335-44. Oncogene. 1994. PMID: 8036016
-
Understanding Cell Lines, Patient-Derived Xenograft and Genetically Engineered Mouse Models Used to Study Cutaneous T-Cell Lymphoma.Cells. 2022 Feb 9;11(4):593. doi: 10.3390/cells11040593. Cells. 2022. PMID: 35203244 Free PMC article. Review.
-
Insights Into the Molecular and Cellular Underpinnings of Cutaneous T Cell Lymphoma.Yale J Biol Med. 2020 Mar 27;93(1):111-121. eCollection 2020 Mar. Yale J Biol Med. 2020. PMID: 32226341 Free PMC article. Review.
Cited by
-
Molecular Landscape of Endometrial Stromal Tumors.JCO Precis Oncol. 2025 May;9:e2400779. doi: 10.1200/PO-24-00779. Epub 2025 May 22. JCO Precis Oncol. 2025. PMID: 40403211 Free PMC article.
References
-
- Vermeer M.H., van Doorn R., Dijkman R., Mao X., Whittaker S., van Voorst Vader P.C., Gerritsen M.J.P., Geerts M.L., Gellrich S., Soderberg O., et al. Novel and highly recurrent chromosomal alterations in Sezary syndrome. Cancer Res. 2008;68:2689–2698. doi: 10.1158/0008-5472.CAN-07-6398. - DOI - PubMed
-
- Laharanne E., Oumouhou N., Bonnet F., Carlotti M., Gentil C., Chevret E., Jouary T., Longy M., Vergier B., Beylot-Barry M., et al. Genome-wide analysis of cutaneous T-cell lymphomas identifies three clinically relevant classes. J. Investig. Dermatol. 2010;130:1707–1718. doi: 10.1038/jid.2010.8. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

