Transcriptional Regulator DasR Represses Daptomycin Production through Both Direct and Cascade Mechanisms in Streptomyces roseosporus
- PMID: 36009934
- PMCID: PMC9404778
- DOI: 10.3390/antibiotics11081065
Transcriptional Regulator DasR Represses Daptomycin Production through Both Direct and Cascade Mechanisms in Streptomyces roseosporus
Abstract
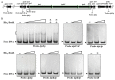
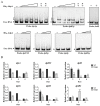
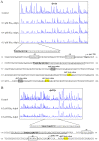
Daptomycin, produced by Streptomyces roseosporus, is a clinically important cyclic lipopeptide antibiotic used for the treatment of human infections caused by drug-resistant Gram-positive pathogens. In contrast to most Streptomyces antibiotic biosynthetic gene clusters (BGCs), daptomycin BGC has no cluster-situated regulator (CSR) genes. DasR, a GntR-family transcriptional regulator (TR) widely present in the genus, was shown to regulate antibiotic production in model species S. coelicolor by binding to promoter regions of CSR genes. New findings reported here reveal that DasR pleiotropically regulates production of daptomycin and reddish pigment, and morphological development in S. roseosporus. dasR deletion enhanced daptomycin production and morphological development, but reduced pigment production. DasR inhibited daptomycin production by directly repressing dpt structural genes and global regulatory gene adpA (whose product AdpA protein activates daptomycin production and morphological development). DasR-protected regions on dptEp and adpAp contained a 16 nt sequence similar to the consensus DasR-binding site dre in S. coelicolor. AdpA was shown to target dpt structural genes and dptR2 (which encodes a DeoR-family TR required for daptomycin production). A 10 nt sequence similar to the consensus AdpA-binding site was found on target promoter regions dptAp and dptR2p. This is the first demonstration that DasR regulates antibiotic production both directly and through a cascade mechanism. The findings expand our limited knowledge of the regulatory network underlying daptomycin production, and will facilitate methods for construction of daptomycin overproducers.
Keywords: AdpA; DasR; Streptomyces roseosporus; daptomycin; morphological development.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures






References
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

