Queuine Salvaging in the Human Parasite Entamoeba histolytica
- PMID: 36010587
- PMCID: PMC9406330
- DOI: 10.3390/cells11162509
Queuine Salvaging in the Human Parasite Entamoeba histolytica
Abstract
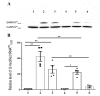
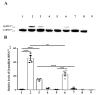
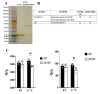
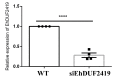
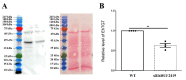
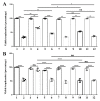
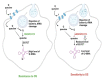
Queuosine (Q) is a naturally occurring modified nucleoside that occurs in the first position of transfer RNA anticodons such as Asp, Asn, His, and Tyr. As eukaryotes lack pathways to synthesize queuine, the Q nucleobase, they must obtain it from their diet or gut microbiota. Previously, we described the effects of queuine on the physiology of the eukaryotic parasite Entamoeba histolytica and characterized the enzyme EhTGT responsible for queuine incorporation into tRNA. At present, it is unknown how E. histolytica salvages queuine from gut bacteria. We used liquid chromatography-mass spectrometry (LC-MS) and N-acryloyl-3-aminophenylboronic acid (APB) PAGE analysis to demonstrate that E. histolytica trophozoites can salvage queuine from Q or E. coli K12 but not from the modified E. coli QueC strain, which cannot produce queuine. We then examined the role of EhDUF2419, a protein with homology to DNA glycosylase, as a queuine salvage enzyme in E. histolytica. We found that glutathione S-transferase (GST)-EhDUF2419 catalyzed the conversion of Q into queuine. Trophozoites silenced for EhDUF2419 expression are impaired in their ability to form Q-tRNA from Q or from E. coli. We also observed that Q or E. coli K12 partially protects control trophozoites from oxidative stress (OS), but not siEhDUF2419 trophozoites. Overall, our data reveal that EhDUF2419 is central for the direct salvaging of queuine from bacteria and for the resistance of the parasite to OS.
Keywords: Entamoeba histolytica; oxidative stress resistance; queuine; queuosine; tRNA modifications.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

