Elicitation of Roots and AC-DC with PEP-13 Peptide Shows Differential Defense Responses in Multi-Omics
- PMID: 36010682
- PMCID: PMC9406913
- DOI: 10.3390/cells11162605
Elicitation of Roots and AC-DC with PEP-13 Peptide Shows Differential Defense Responses in Multi-Omics
Abstract
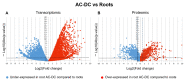
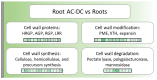
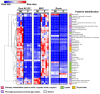
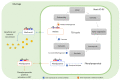
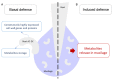
The root extracellular trap (RET) has emerged as a specialized compartment consisting of root AC-DC and mucilage. However, the RET's contribution to plant defense is still poorly understood. While the roles of polysaccharides and glycoproteins secreted by root AC-DC have started to be elucidated, how the low-molecular-weight exudates of the RET contribute to root defense is poorly known. In order to better understand the RET and its defense response, the transcriptomes, proteomes and metabolomes of roots, root AC-DC and mucilage of soybean (Glycine max (L.) Merr, var. Castetis) upon elicitation with the peptide PEP-13 were investigated. This peptide is derived from the pathogenic oomycete Phytophthora sojae. In this study, the root and the RET responses to elicitation were dissected and sequenced using transcriptional, proteomic and metabolomic approaches. The major finding is increased synthesis and secretion of specialized metabolites upon induced defense activation following PEP-13 peptide elicitation. This study provides novel findings related to the pivotal role of the root extracellular trap in root defense.
Keywords: Glycine max; PEP-13 elicitor; omics; root-associated cap-derived cells (root AC-DC).
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Hawes M.C., Bengough G., Cassab G., Ponce G. Root Caps and Rhizosphere. J. Plant Growth Regul. 2002;21:352–367. doi: 10.1007/s00344-002-0035-y. - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

