Estrogen Related Receptor Alpha (ERRα) a Bridge between Metabolism and Adrenocortical Cancer Progression
- PMID: 36010877
- PMCID: PMC9406166
- DOI: 10.3390/cancers14163885
Estrogen Related Receptor Alpha (ERRα) a Bridge between Metabolism and Adrenocortical Cancer Progression
Abstract
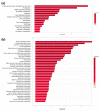
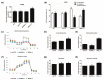
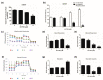
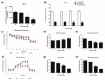
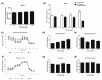
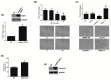
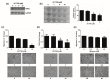
The aim of this study was to investigate the metabolic changes that occur in adrenocortical cancer (ACC) cells in response to the modulation of Estrogen Related Receptor (ERR)α expression and the impact on ACC progression. Proteomics analysis and metabolic profiling highlighted an important role for ERRα in the regulation of ACC metabolism. Stable ERRα overexpression in H295R cells promoted a better mitochondrial fitness and prompted toward a more aggressive phenotype characterized by higher Vimentin expression, enhanced cell migration and spheroids formation. By contrast, a decrease in ERRα protein levels, by molecular (short hairpin RNA) and pharmacological (inverse agonist XCT790) approaches modified the energetic status toward a low energy profile and reduced Vimentin expression and ability to form spheroids. XCT790 produced similar effects on two additional ACC cell lines, SW13 and mitotane-resistant MUC-1 cells. Our findings show that ERRα is able to modulate the metabolic profile of ACC cells, and its inhibition can strongly prevent the growth of mitotane-resistant ACC cells and the progression of ACC cell models to a highly migratory phenotype. Consequently, ERRα can be considered an important target for the design of new therapeutic strategies to fight ACC progression.
Keywords: ERRα; XCT790; adrenocortical cancer; cancer progression; metabolic changes; mitochondria.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Vatrano S., Volante M., Duregon E., Giorcelli J., Izzo S., Rapa I., Votta A., Germano A., Scagliotti G., Berruti A., et al. Detailed genomic characterization identifies high heterogeneity and histotype-specific genomic profiles in adrenocortical carcinomas. Mod. Pathol. 2018;31:1257–1269. doi: 10.1038/s41379-018-0042-6. - DOI - PubMed
Grants and funding
- Italian Law 232/2016/Special award (Department of Excellence, Italian Law 232/2016) from the Italian Ministry of Research and University (MIUR) to the Department of Pharmacy, Health and Nutritional Sciences of the University of Calabria (Italy),
- projects n. IG20122/Italian Association for Cancer Research
- ex 60%/Ministry of Education, Universities and Research
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

