The Methylation of the p53 Targets the Genes MIR-203, MIR-129-2, MIR-34A and MIR-34B/C in the Tumor Tissue of Diffuse Large B-Cell Lymphoma
- PMID: 36011313
- PMCID: PMC9408007
- DOI: 10.3390/genes13081401
The Methylation of the p53 Targets the Genes MIR-203, MIR-129-2, MIR-34A and MIR-34B/C in the Tumor Tissue of Diffuse Large B-Cell Lymphoma
Abstract
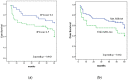
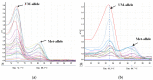
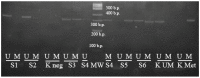
The regulation of oncogenes by microRNA is a focus of medical research. hsa-miR-203, hsa-mir-129, hsa-miR-34a, hsa-miR-34b and hsa-miR-34c are oncosuppressive microRNAs that mediate the antitumor activity of p53. We seek to evaluate the frequencies, co-occurrence and clinical significance of the methylation of the MIR-203, MIR-129-2, MIR-34A and MIR-34B/C genes in the tumor tissue of diffuse large B-cell lymphoma (DLBCL). The methylation was assessed in 73 samples of DLBCL and in 11 samples of lymph nodes of reactive follicular hyperplasia by Methyl-Specific Polymerase Chain Reaction (MS-PCR) and Methylation-Sensitive High-Resolution-Melting (MS-HRM) methods. All four studied genes were not methylated in the tissue of reactive lymphatic nodes. The methylation frequencies of the MIR-129-2, MIR-203, MIR-34A and MIR-34B/C genes in lymphoma tissue were 67%, 66%, 27% and 62%, respectively. Co-occurrence of MIR-203, MIR-129-2 and MIR-34B/C genes methylation, as well as the methylation of MIR-34B/C and MIR-34A pair genes were detected. The MIR-34A gene methylation was associated with increased International Prognostic Index (IPI) (p = 0.002), whereas the MIR-34B/C (p = 0.026) and MIR-203 (p = 0.011) genes' methylation was connected with Ki-67 expression level in tumor tissue at more than 45%. We found an increasing frequency of detection of MIR-34A gene methylation in the group of patients with the Germinal-Center B-cell like (GCB-like) subtype of DLBCL (p = 0.046). There was a trend towards a decrease in the remission frequency after the first line of therapy (p = 0.060) and deterioration in overall survival (OS) (p = 0.162) in patients with DLBCL with methylation of the MIR-34A promoter. The methylation of the MIR-34A, MIR-34B/C, MIR-129-2 and MIR-203 genes in DLBCL is tumor-specific and occurs in combination. The methylation of the studied genes may be a potential differential diagnostic biomarker to distinguish between lymphoma and reactive lymph nodes, while its independent predictive value has not been confirmed yet.
Keywords: diffuse large B-cell lymphoma; methylation; microRNA.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Aushev V.N. MicroRNA: Small Molecules of Great Significance. Clin. Oncohematology. 2015;8:1–12. (In Russian)
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

