Molecular Dynamic Simulation Reveals Structure Differences in APOL1 Variants and Implication in Pathogenesis of Chronic Kidney Disease
- PMID: 36011371
- PMCID: PMC9408642
- DOI: 10.3390/genes13081460
Molecular Dynamic Simulation Reveals Structure Differences in APOL1 Variants and Implication in Pathogenesis of Chronic Kidney Disease
Abstract
Background: According to observational studies, two polymorphisms in the apolipoprotein L1 (APOL1) gene have been linked to an increased risk of chronic kidney disease (CKD) in Africans. One polymorphism involves the substitution of two amino-acid residues (S342G and I384M; known as G1), while the other involves the deletion of two amino-acid residues in a row (N388 and Y389; termed G2). Despite the strong link between APOL1 polymorphisms and kidney disease, the molecular mechanisms via which these APOL1 mutations influence the onset and progression of CKD remain unknown.
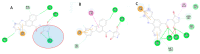
Methods: To predict the active site and allosteric site on the APOL1 protein, we used the Computed Atlas of Surface Topography of Proteins (CASTp) and the Protein Allosteric Sites Server (PASSer). Using an extended molecular dynamics simulation, we investigated the characteristic structural perturbations in the 3D structures of APOL1 variants.
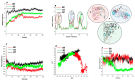
Results: According to CASTp's active site characterization, the topmost predicted site had a surface area of 964.892 Å2 and a pocket volume of 900.792 Å3. For the top three allosteric pockets, the allostery probability was 52.44%, 46.30%, and 38.50%, respectively. The systems reached equilibrium in about 125 ns. From 0-100 ns, there was also significant structural instability. When compared to G1 and G2, the wildtype protein (G0) had overall high stability throughout the simulation. The root-mean-square fluctuation (RMSF) of wildtype and variant protein backbone Cα fluctuations revealed that the Cα of the variants had a large structural fluctuation when compared to the wildtype.
Conclusion: Using a combination of different computational techniques, we identified binding sites within the APOL1 protein that could be an attractive site for potential inhibitors of APOL1. Furthermore, the G1 and G2 mutations reduced the structural stability of APOL1.
Keywords: APOL1; chronic kidney disease; molecular docking; molecular dynamic simulation; mutation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Kidney Disease Statistics for the United States|NIDDK. [(accessed on 7 March 2022)]; Available online: https://www.niddk.nih.gov/health-information/health-statistics/kidney-di....
-
- Kidney Disease/Chronic Kidney Disease: Symptoms, Treatment & More. [(accessed on 7 March 2022)]. Available online: https://my.clevelandclinic.org/health/diseases/15096-kidney-disease-chro....
-
- Things to Know About Kidney Function|National Kidney Foundation. [(accessed on 7 March 2022)]. Available online: https://www.kidney.org/kidneydisease/howkidneyswrk.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

