CDC42-IQGAP Interactions Scrutinized: New Insights into the Binding Properties of the GAP-Related Domain
- PMID: 36012107
- PMCID: PMC9408373
- DOI: 10.3390/ijms23168842
CDC42-IQGAP Interactions Scrutinized: New Insights into the Binding Properties of the GAP-Related Domain
Abstract
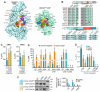
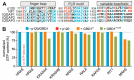
The IQ motif-containing GTPase-activating protein (IQGAP) family composes of three highly-related and evolutionarily conserved paralogs (IQGAP1, IQGAP2 and IQGAP3), which fine tune as scaffolding proteins numerous fundamental cellular processes. IQGAP1 is described as an effector of CDC42, although its effector function yet re-mains unclear. Biophysical, biochemical and molecular dynamic simulation studies have proposed that IQGAP RASGAP-related domains (GRDs) bind to the switch regions and the insert helix of CDC42 in a GTP-dependent manner. Our kinetic and equilibrium studies have shown that IQGAP1 GRD binds, in contrast to its C-terminal 794 amino acids (called C794), CDC42 in a nucleotide-independent manner indicating a binding outside the switch regions. To resolve this discrepancy and move beyond the one-sided view of GRD, we carried out affinity measurements and a systematic mutational analysis of the interfacing residues between GRD and CDC42 based on the crystal structure of the IQGAP2 GRD-CDC42Q61L GTP complex. We determined a 100-fold lower affinity of the GRD1 of IQGAP1 and of GRD2 of IQGAP2 for CDC42 mGppNHp in comparison to C794/C795 proteins. Moreover, partial and major mutation of CDC42 switch regions substantially affected C794/C795 binding but only a little GRD1 and remarkably not at all the GRD2 binding. However, we clearly showed that GRD2 contributes to the overall affinity of C795 by using a 11 amino acid mutated GRD variant. Furthermore, the GRD1 binding to the CDC42 was abolished using specific point mutations within the insert helix of CDC42 clearly supporting the notion that CDC42 binding site(s) of IQGAP GRD lies outside the switch regions among others in the insert helix. Collectively, this study provides further evidence for a mechanistic framework model that is based on a multi-step binding process, in which IQGAP GRD might act as a 'scaffolding domain' by binding CDC42 irrespective of its nucleotide-bound forms, followed by other IQGAP domains downstream of GRD that act as an effector domain and is in charge for a GTP-dependent interaction with CDC42.
Keywords: CDC42; GAP; GAP-related domain; GRD; GTPase activating protein; IQGAP; RASGAP; RHO GTPases; nucleotide-independent binding; scaffold protein; scaffolding protein; switch regions.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Unraveling the molecular mechanism of interactions of the Rho GTPases Cdc42 and Rac1 with the scaffolding protein IQGAP2.J Biol Chem. 2018 Mar 9;293(10):3685-3699. doi: 10.1074/jbc.RA117.001596. Epub 2018 Jan 22. J Biol Chem. 2018. PMID: 29358323 Free PMC article.
-
The Structural Basis for Cdc42-Induced Dimerization of IQGAPs.Structure. 2016 Sep 6;24(9):1499-508. doi: 10.1016/j.str.2016.06.016. Epub 2016 Aug 11. Structure. 2016. PMID: 27524202 Free PMC article.
-
IQGAP1 Interaction with RHO Family Proteins Revisited: KINETIC AND EQUILIBRIUM EVIDENCE FOR MULTIPLE DISTINCT BINDING SITES.J Biol Chem. 2016 Dec 16;291(51):26364-26376. doi: 10.1074/jbc.M116.752121. Epub 2016 Nov 4. J Biol Chem. 2016. PMID: 27815503 Free PMC article.
-
New model for the interaction of IQGAP1 with CDC42 and RAC1.Small GTPases. 2020 Jan;11(1):16-22. doi: 10.1080/21541248.2017.1321169. Epub 2017 Jun 19. Small GTPases. 2020. PMID: 28622072 Free PMC article. Review.
-
IQGAPs in cancer: a family of scaffold proteins underlying tumorigenesis.FEBS Lett. 2009 Jun 18;583(12):1817-24. doi: 10.1016/j.febslet.2009.05.007. Epub 2009 May 9. FEBS Lett. 2009. PMID: 19433088 Free PMC article. Review.
Cited by
-
The IQGAP-related RasGAP IqgC regulates cell-substratum adhesion in Dictyostelium discoideum.Cell Mol Biol Lett. 2025 Jan 9;30(1):4. doi: 10.1186/s11658-024-00678-3. Cell Mol Biol Lett. 2025. PMID: 39789437 Free PMC article.
-
Modulating PAK1: Accessory Proteins as Promising Therapeutic Targets.Biomolecules. 2025 Feb 7;15(2):242. doi: 10.3390/biom15020242. Biomolecules. 2025. PMID: 40001545 Free PMC article. Review.
-
Loss of SIL1 Affects Actin Dynamics and Leads to Abnormal Neural Migration.Mol Neurobiol. 2025 Jan;62(1):335-350. doi: 10.1007/s12035-024-04272-8. Epub 2024 Jun 8. Mol Neurobiol. 2025. PMID: 38850350
References
MeSH terms
Substances
Grants and funding
- 01GM1621B/the European Network on Noonan Syndrome and Related Disorders (NSEuroNet)
- 01GM1902C/Federal Ministry of Education and Research
- IRTG 1902-p6/Deutsche Forschungsgemeinschaft
- 9772764/the Research Committee of the Medical Faculty of the Heinrich Heine University
- ITMS2014+: 313011V455/the Operational Program Integrated Infrastructure ERDF: Open scientific community for modern interdisciplinary research in medicine (OPENMED)
LinkOut - more resources
Full Text Sources
Miscellaneous

