A Comparative Study of the Plasma Chemokine Profile in COVID-19 Patients Infected with Different SARS-CoV-2 Variants
- PMID: 36012323
- PMCID: PMC9409001
- DOI: 10.3390/ijms23169058
A Comparative Study of the Plasma Chemokine Profile in COVID-19 Patients Infected with Different SARS-CoV-2 Variants
Abstract
Background: Infection caused by SARS-CoV-2 mostly affects the upper and lower respiratory tracts and causes symptoms ranging from the common cold to pneumonia with acute respiratory distress syndrome. Chemokines are deeply involved in the chemoattraction, proliferation, and activation of immune cells within inflammation. It is crucial to consider that mutations within the virion can potentially affect the clinical course of SARS-CoV-2 infection because disease severity and manifestation vary depending on the genetic variant. Our objective was to measure and assess the different concentrations of chemokines involved in COVID-19 caused by different variants of the virus.
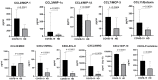
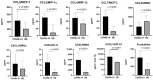
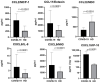
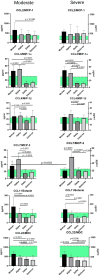
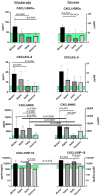
Methods: We used the blood plasma of patients infected with different variants of SARS-CoV-2, i.e., the ancestral Wuhan strain and the Alpha, Delta, and Omicron variants. We measured the concentrations of 11 chemokines in the samples: CCL2/MCP-1, CCL3/MIP-1α, CCL4/MIP-1β, CCL7/MCP-3, CCL11/Eotaxin, CCL22/MDC, CXCL1/GROα, CXCL8/IL-8, CXCL9/MIG, CXCL10/IP-10, and CX3CL1/Fractalkine.
Results: We noted a statistically significant elevation in the concentrations of CCL2/MCP-1, CXCL8/IL-8, and CXCL1/IP-10 independently of the variant, and a drop in the CCL22/MDC concentrations.
Conclusions: The chemokine concentrations varied significantly depending on the viral variant, leading us to infer that mutations in viral proteins play a role in the cellular and molecular mechanisms of immune responses.
Keywords: COVID-19; Omicron; SARS-CoV-2 variants; chemokines; macrophage-derived chemokine.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- D’Agnillo F., Walters K.-A., Xiao Y., Sheng Z.-M., Scherler K., Park J., Gygli S., Rosas L.A., Sadtler K., Kalish H., et al. Lung epithelial and endothelial damage, loss of tissue repair, inhibition of fibrinolysis, and cellular senescence in fatal COVID-19. Sci. Transl. Med. 2021;13:eabj7790. doi: 10.1126/scitranslmed.abj7790. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

