High IGKC-Expressing Intratumoral Plasma Cells Predict Response to Immune Checkpoint Blockade
- PMID: 36012390
- PMCID: PMC9408876
- DOI: 10.3390/ijms23169124
High IGKC-Expressing Intratumoral Plasma Cells Predict Response to Immune Checkpoint Blockade
Abstract
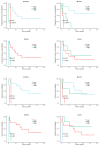
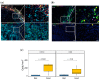
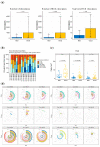
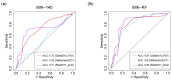
Resistance to Immune Checkpoint Blockade (ICB) constitutes the current limiting factor for the optimal implementation of this novel therapy, which otherwise demonstrates durable responses with acceptable toxicity scores. This limitation is exacerbated by a lack of robust biomarkers. In this study, we have dissected the basal TME composition at the gene expression and cellular levels that predict response to Nivolumab and prognosis. BCR, TCR and HLA profiling were employed for further characterization of the molecular variables associated with response. The findings were validated using a single-cell RNA-seq data of metastatic melanoma patients treated with ICB, and by multispectral immunofluorescence. Finally, machine learning was employed to construct a prediction algorithm that was validated across eight metastatic melanoma cohorts treated with ICB. Using this strategy, we have unmasked a major role played by basal intratumoral Plasma cells expressing high levels of IGKC in efficacy. IGKC, differentially expressed in good responders, was also identified within the Top response-related BCR clonotypes, together with IGK variants. These results were validated at gene, cellular and protein levels; CD138+ Plasma-like and Plasma cells were more abundant in good responders and correlated with the same RNA-seq-defined fraction. Finally, we generated a 15-gene prediction model that outperformed the current reference score in eight ICB-treated metastatic melanoma cohorts. The evidenced major contribution of basal intratumoral IGKC and Plasma cells in good response and outcome in ICB in metastatic melanoma is a groundbreaking finding in the field beyond the role of T lymphocytes.
Keywords: biomarkers; immunotherapy; melanoma.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Ribas A., Kefford R., Marshall M.A., Punt C.J.A., Haanen J.B., Marmol M., Garbe C., Gogas H., Schachter J., Linette G., et al. Phase III Randomized Clinical Trial Comparing Tremelimumab with Standard-of-Care Chemotherapy in Patients with Advanced Melanoma. J. Clin. Oncol. 2013;31:616–622. doi: 10.1200/JCO.2012.44.6112. - DOI - PMC - PubMed
-
- Ribas A., Puzanov I., Dummer R., Schadendorf D., Hamid O., Robert C., Hodi F.S., Schachter J., Pavlick A.C., Lewis K.D., et al. Pembrolizumab versus Investigator-Choice Chemotherapy for Ipilimumab-Refractory Melanoma (KEYNOTE-002): A Randomised, Controlled, Phase 2 Trial. Lancet Oncol. 2015;16:908–918. doi: 10.1016/S1470-2045(15)00083-2. - DOI - PMC - PubMed
-
- Mok T.S.K., Wu Y.L., Kudaba I., Kowalski D.M., Cho B.C., Turna H.Z., Castro G., Srimuninnimit V., Laktionov K.K., Bondarenko I., et al. Pembrolizumab versus Chemotherapy for Previously Untreated, PD-L1-Expressing, Locally Advanced or Metastatic Non-Small-Cell Lung Cancer (KEYNOTE-042): A Randomised, Open-Label, Controlled, Phase 3 Trial. Lancet. 2019;393:1819–1830. doi: 10.1016/S0140-6736(18)32409-7. - DOI - PubMed
MeSH terms
Substances
Grants and funding
- PI18/01592/Instituto de Salud Carlos III
- FI19-00112/Instituto de Salud Carlos III
- SA 0263/2017/Sistema Andaluz de Salud
- Nicolás Monardes/Sistema Andaluz de Salud
- PI-0121-2020/Sistema Andaluz de Salud
- RH-0090-2020/Sistema Andaluz de Salud
- C19048/Fundación Bancaria Unicaja
- Precision Medical Oncology/Andalusia-Roche Network Mixed Alliance
- PI-0121-2020/Consejería de Salud y Familias
- CV20-62050/Consejería de Transformación Económica, Industria, Conocimiento y Ciencia
- 201600160066/China Scholarship Council
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

