Stabilization of the c-Myc Protein via the Modulation of Threonine 58 and Serine 62 Phosphorylation by the Disulfiram/Copper Complex in Oral Cancer Cells
- PMID: 36012403
- PMCID: PMC9409128
- DOI: 10.3390/ijms23169137
Stabilization of the c-Myc Protein via the Modulation of Threonine 58 and Serine 62 Phosphorylation by the Disulfiram/Copper Complex in Oral Cancer Cells
Abstract
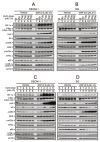
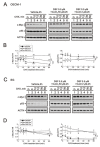
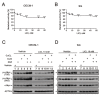
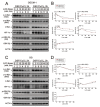
MYC has a short half-life that is tightly regulated through phosphorylation and proteasomal degradation. Many studies have claimed that treatment with disulfiram (DSF) with or without copper ions can cause cancer cell death in a reactive oxygen species (ROS)-dependent manner in cancer cells. Our previous study showed that the levels of c-Myc protein and the phosphorylation of threonine 58 (T58) and serine 62 (S62) increased in DSF-Cu-complex-treated oral epidermoid carcinoma Meng-1 (OECM-1) cells. These abovementioned patterns were suppressed by pretreatment with an ROS scavenger, N-acetyl cysteine. The overexpression of c-Myc failed to induce hypoxia-inducible factor 1α protein expression, which was stabilized by the DSF-Cu complex. In this study, we further examined the regulatory mechanism behind the induction of the c-Myc of the DSF-Cu complex in an OECM-1 cell compared with a Smulow-Glickman (SG) human normal gingival epithelial cell. Our data showed that the downregulation of c-Myc truncated nick and p62 and the induction of the ratio of H3P/H3 and p-ERK/ERK might not be involved in the increase in the amount of c-Myc via the DSF/copper complexes in OECM-1 cells. Combined with the inhibitors for various signaling pathways and cycloheximde treatment, the increase in the amount of c-Myc with the DSF/copper complexes might be mediated through the increase in the stabilities of c-Myc (T58) and c-Myc (S62) proteins in OECM-1 cells. In SG cells, only the c-Myc (T58) protein was stabilized by the DSF-Cu (I and II) complexes. Hence, our findings could provide novel regulatory insights into the phosphorylation-dependent stability of c-Myc in DSF/copper-complex-treated oral squamous cell carcinoma.
Keywords: c-Myc; disulfiram; hypoxia inducible factor 1α; reactive oxygen species.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

