Plasma Microbiome in COVID-19 Subjects: An Indicator of Gut Barrier Defects and Dysbiosis
- PMID: 36012406
- PMCID: PMC9409329
- DOI: 10.3390/ijms23169141
Plasma Microbiome in COVID-19 Subjects: An Indicator of Gut Barrier Defects and Dysbiosis
Abstract
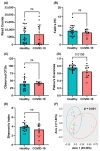
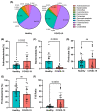
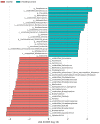
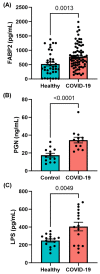
The gut is a well-established route of infection and target for viral damage by SARS-CoV-2. This is supported by the clinical observation that about half of COVID-19 patients exhibit gastrointestinal (GI) complications. We aimed to investigate whether the analysis of plasma could provide insight into gut barrier dysfunction in patients with COVID-19 infection. Plasma samples of COVID-19 patients (n = 146) and healthy individuals (n = 47) were collected during hospitalization and routine visits. Plasma microbiome was analyzed using 16S rRNA sequencing and gut permeability markers including fatty acid binding protein 2 (FABP2), peptidoglycan (PGN), and lipopolysaccharide (LPS) in both patient cohorts. Plasma samples of both cohorts contained predominately Proteobacteria, Firmicutes, Bacteroides, and Actinobacteria. COVID-19 subjects exhibit significant dysbiosis (p = 0.001) of the plasma microbiome with increased abundance of Actinobacteria spp. (p = 0.0332), decreased abundance of Bacteroides spp. (p = 0.0003), and an increased Firmicutes:Bacteroidetes ratio (p = 0.0003) compared to healthy subjects. The concentration of the plasma gut permeability marker FABP2 (p = 0.0013) and the gut microbial antigens PGN (p < 0.0001) and LPS (p = 0.0049) were significantly elevated in COVID-19 patients compared to healthy subjects. These findings support the notion that the intestine may represent a source for bacteremia and contribute to worsening COVID-19 outcomes. Therapies targeting the gut and prevention of gut barrier defects may represent a strategy to improve outcomes in COVID-19 patients.
Keywords: COVID-19; circulating microbiome; dysbiosis; gut barrier permeability.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Update of
-
Plasma microbiome in COVID-19 subjects: an indicator of gut barrier defects and dysbiosis.bioRxiv [Preprint]. 2021 Apr 6:2021.04.06.438634. doi: 10.1101/2021.04.06.438634. bioRxiv. 2021. Update in: Int J Mol Sci. 2022 Aug 15;23(16):9141. doi: 10.3390/ijms23169141. PMID: 33851159 Free PMC article. Updated. Preprint.
References
-
- WHO Coronavirus (COVID-19) Dashboard. [(accessed on 21 July 2022)]. Available online: https://covid19.who.int/
-
- COVID-19 Weekly Cases and Deaths per 100,000 Population by Age, Race/Ethnicity, and Sex. [(accessed on 21 July 2022)]; Available online: https://covid.cdc.gov/covid-data-tracker/#demographicsovertime.
-
- Guzik T.J., Mohiddin S.A., Dimarco A., Patel V., Savvatis K., Marelli-Berg F.M., Madhur M.S., Tomaszewski M., Maffia P., D’Acquisto F., et al. COVID-19 and the cardiovascular system: Implications for risk assessment, diagnosis, and treatment options. Cardiovasc. Res. 2020;116:1666–1687. doi: 10.1093/cvr/cvaa106. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

