Allelic Variations in the Human Genes TMPRSS2 and CCR5, and the Resistance to Viral Infection by SARS-CoV-2
- PMID: 36012436
- PMCID: PMC9409186
- DOI: 10.3390/ijms23169171
Allelic Variations in the Human Genes TMPRSS2 and CCR5, and the Resistance to Viral Infection by SARS-CoV-2
Abstract
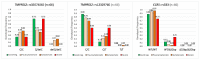
During the first wave of COVID-19 infection in Italy, the number of cases and the mortality rates were among the highest compared to the rest of Europe and the world. Several studies demonstrated a severe clinical course of COVID-19 associated with old age, comorbidities, and male gender. However, there are cases of virus infection resistance in subjects living in close contact with infected subjects. Thus, to explain the predisposition to virus infection and to COVID-19 disease progression, we must consider, in addition to the genetic variability of the virus and other environmental or comorbidity conditions, the allelic variants of specific human genes, directly or indirectly related to the life cycle of the virus. Here, we analyzed three human genetic polymorphisms belonging to the TMPRSS2 and CCR5 genes in a sample population from Sicily (Italy) to investigate possible correlations with the resistance to viral infection and/or to COVID-19 disease progression as recently described in other human populations. Our results did not show any correlations of the rs35074065, rs12329760, and rs333 polymorphisms with SARS-CoV-2 infection or with COVID-19 disease severity. Further studies on other human genetic polymorphisms should be performed to identify the major human determinants of SARS-CoV-2 viral resistance.
Keywords: CCR5 gene; COVID-19; SARS-CoV-2 first wave infection; TMPRSS2 gene; rs12329760; rs333; rs35074065.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Sedlmaier N., Hoppenheidt K., Krist H., Lehmann S., Lang H., Büttner M. Generation of avian influenza virus (AIV) contaminated fecal fine particulate matter (PM(2.5): Genome and infectivity detection and calculation of immission. Vet. Microbiol. 2009;139:156–164. doi: 10.1016/j.vetmic.2009.05.005. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

