Identification and Characterization of Jasmonic Acid Biosynthetic Genes in Salvia miltiorrhiza Bunge
- PMID: 36012649
- PMCID: PMC9409215
- DOI: 10.3390/ijms23169384
Identification and Characterization of Jasmonic Acid Biosynthetic Genes in Salvia miltiorrhiza Bunge
Abstract
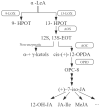
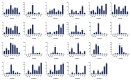
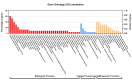
Jasmonic acid (JA) is a vital plant hormone that performs a variety of critical functions for plants. Salvia miltiorrhiza Bunge (S. miltiorrhiza), also known as Danshen, is a renowned traditional Chinese medicinal herb. However, no thorough and systematic analysis of JA biosynthesis genes in S. miltiorrhiza exists. Through genome-wide prediction and molecular cloning, 23 candidate genes related to JA biosynthesis were identified in S. miltiorrhiza. These genes belong to four families that encode lipoxygenase (LOX), allene oxide synthase (AOS), allene oxide cyclase (AOC), and 12-OPDA reductase3 (OPR3). It was discovered that the candidate genes for JA synthesis of S. miltiorrhiza were distinct and conserved, in contrast to related genes in other plants, by evaluating their genetic structures, protein characteristics, and phylogenetic trees. These genes displayed tissue-specific expression patterns concerning to methyl jasmonate (MeJA) and wound tests. Overall, the results of this study provide valuable information for elucidating the JA biosynthesis pathway in S. miltiorrhiza by comprehensive and methodical examination.
Keywords: Salvia miltiorrhiza; jasmonic acid; secondary metabolism; traditional Chinese medicine.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

