Selective Expansion of NKG2C+ Adaptive NK Cells Using K562 Cells Expressing HLA-E
- PMID: 36012691
- PMCID: PMC9409060
- DOI: 10.3390/ijms23169426
Selective Expansion of NKG2C+ Adaptive NK Cells Using K562 Cells Expressing HLA-E
Abstract
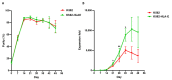
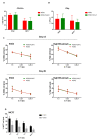
Adaptive natural killer (NK) cells expressing self-specific inhibitory killer-cell immunoglobulin-like receptors (KIRs) can be expanded in vivo in response to human cytomegalovirus (HCMV) infection. Developing a method to preferentially expand this subset is essential for effective targeting of allogeneic cancer cells. A previous study developed an in vitro method to generate single KIR+ NK cells for enhanced targeting of the primary acute lymphoblastic leukemia cells; however, the expansion rate was quite low. Here, we present an effective expansion method using genetically modified K562-HLA-E feeder cells for long-term proliferation of adaptive NK cells displaying highly differentiated phenotype and comparable cytotoxicity, CD107a, and interferon-γ (IFN-γ) production. More importantly, our expansion method achieved more than a 10,000-fold expansion of adaptive NK cells after 6 weeks of culture, providing a high yield of alloreactive NK cells for cell therapy against cancer.
Keywords: K562-HLA-E feeder cells; adaptive NK cells; effective expansion; long-term persistence; single KIR NKG2C.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as potential conflicts of interest.
Figures




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

