Artificial Intelligence in Cryo-Electron Microscopy
- PMID: 36013446
- PMCID: PMC9410485
- DOI: 10.3390/life12081267
Artificial Intelligence in Cryo-Electron Microscopy
Abstract
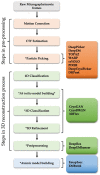
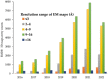
Cryo-electron microscopy (cryo-EM) has become an unrivaled tool for determining the structure of macromolecular complexes. The biological function of macromolecular complexes is inextricably tied to the flexibility of these complexes. Single particle cryo-EM can reveal the conformational heterogeneity of a biochemically pure sample, leading to well-founded mechanistic hypotheses about the roles these complexes play in biology. However, the processing of increasingly large, complex datasets using traditional data processing strategies is exceedingly expensive in both user time and computational resources. Current innovations in data processing capitalize on artificial intelligence (AI) to improve the efficiency of data analysis and validation. Here, we review new tools that use AI to automate the data analysis steps of particle picking, 3D map reconstruction, and local resolution determination. We discuss how the application of AI moves the field forward, and what obstacles remain. We also introduce potential future applications of AI to use cryo-EM in understanding protein communities in cells.
Keywords: artificial intelligence; cryo-electron microscopy; deep learning; neural network.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Chung J.M., Jung H.S. Cryo-electron tomography: A tool for in situ structural analysis of macromolecular complexes. Appl. Spectrosc. Rev. 2017;53:195–202. doi: 10.1080/05704928.2017.1328426. - DOI
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources

