Microbial Biofilms at Meat-Processing Plant as Possible Places of Bacteria Survival
- PMID: 36014001
- PMCID: PMC9415349
- DOI: 10.3390/microorganisms10081583
Microbial Biofilms at Meat-Processing Plant as Possible Places of Bacteria Survival
Abstract
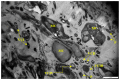
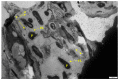
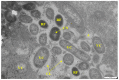
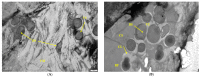
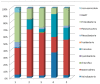
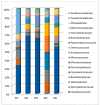
Biofilm contamination in food production threatens food quality and safety, and causes bacterial infections. Study of food biofilms (BF) is of great importance. The taxonomic composition and structural organization of five foods BF taken in different workshops of a meat-processing plant (Moscow, RF) were studied. Samples were taken from the surface of technological equipment and premises. Metagenomic analysis showed both similarities in the presented microorganisms dominating in different samples, and unique families prevailing on certain objects were noted. The bacteria found belonged to 11 phyla (no archaea). The dominant ones were Actinobacteria, Bacteroidetes, Firmicutes, and Proteobacteria. The greatest diversity was in BFs taken from the cutting table of raw material. Biofilms' bacteria may be the cause of meat, fish and dairy products spoilage possible representatives include Pseudomonas, Flavobacterium, Arcobacter, Vagococcus, Chryseobacterium, Carnobacterium, etc.). Opportunistic human and animal pathogens (possible representatives include Arcobacter, Corynebacterium, Kocuria, etc.) were also found. Electron-microscopic studies of BF thin sections revealed the following: (1) the diversity of cell morphotypes specific to multispecies BFs; (2) morphological similarity of cells in BFs from different samples, micro-colonial growth; (3) age heterogeneity of cells within the same microcolony (vegetative and autolyzed cells, resting forms); (4) heterogeneity of the polymer matrix chemical nature according to ruthenium red staining.
Keywords: foodborne pathogens; meat-processing; microbial biofilm.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Identification of biofilm hotspots in a meat processing environment: Detection of spoilage bacteria in multi-species biofilms.Int J Food Microbiol. 2020 Sep 2;328:108668. doi: 10.1016/j.ijfoodmicro.2020.108668. Epub 2020 May 20. Int J Food Microbiol. 2020. PMID: 32474228
-
Tracking bacteriome variation over time in Listeria monocytogenes-positive foci in food industry.Int J Food Microbiol. 2020 Feb 16;315:108439. doi: 10.1016/j.ijfoodmicro.2019.108439. Epub 2019 Nov 6. Int J Food Microbiol. 2020. PMID: 31710972
-
Bacteria of eleven different species isolated from biofilms in a meat processing environment have diverse biofilm forming abilities.Int J Food Microbiol. 2021 Jul 2;349:109232. doi: 10.1016/j.ijfoodmicro.2021.109232. Epub 2021 May 4. Int J Food Microbiol. 2021. PMID: 34022615
-
Attachment and biofilm formation by foodborne bacteria in meat processing environments: causes, implications, role of bacterial interactions and control by alternative novel methods.Meat Sci. 2014 Jul;97(3):298-309. doi: 10.1016/j.meatsci.2013.05.023. Epub 2013 May 23. Meat Sci. 2014. PMID: 23747091 Review.
-
Potential of N2 Gas Flushing to Hinder Dairy-Associated Biofilm Formation and Extension.Front Microbiol. 2020 Jul 28;11:1675. doi: 10.3389/fmicb.2020.01675. eCollection 2020. Front Microbiol. 2020. PMID: 32849349 Free PMC article. Review.
Cited by
-
Ways of Long-Term Survival of Hydrocarbon-Oxidizing Bacteria in a New Biocomposite Material-Silanol-Humate Gel.Microorganisms. 2023 Apr 27;11(5):1133. doi: 10.3390/microorganisms11051133. Microorganisms. 2023. PMID: 37317107 Free PMC article.
-
Hidden Places for Foodborne Bacterial Pathogens and Novel Approaches to Control Biofilms in the Meat Industry.Foods. 2024 Dec 11;13(24):3994. doi: 10.3390/foods13243994. Foods. 2024. PMID: 39766937 Free PMC article. Review.
-
Evaluation of the biofilm-forming ability and molecular characterization of dairy Bacillus spp. isolates.Front Cell Infect Microbiol. 2023 Jul 31;13:1229460. doi: 10.3389/fcimb.2023.1229460. eCollection 2023. Front Cell Infect Microbiol. 2023. PMID: 37600945 Free PMC article.
-
Microbiome Diversity in Seafood Factories via Next-Generation Sequencing for Food Safety Management System (FSMS) Certifications in Malaysia.Foods. 2025 Apr 26;14(9):1517. doi: 10.3390/foods14091517. Foods. 2025. PMID: 40361602 Free PMC article.
-
Understanding how different surfaces and environmental biofilms found in food processing plants affect the spread of COVID-19.PLoS One. 2023 Jun 7;18(6):e0286659. doi: 10.1371/journal.pone.0286659. eCollection 2023. PLoS One. 2023. PMID: 37285373 Free PMC article.
References
-
- Satpathy S., Sen S.K., Pattanaik S., Raut S. Review on bacterial biofilm: An universal cause of contamination. Biocatal. Agric. Biotechnol. 2016;7:56–66. doi: 10.1016/j.bcab.2016.05.002. - DOI
LinkOut - more resources
Full Text Sources
Miscellaneous

