Characterization of Host-Specific Genes from Pine- and Grass-Associated Species of the Fusarium fujikuroi Species Complex
- PMID: 36014979
- PMCID: PMC9415769
- DOI: 10.3390/pathogens11080858
Characterization of Host-Specific Genes from Pine- and Grass-Associated Species of the Fusarium fujikuroi Species Complex
Abstract
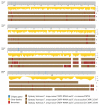
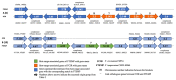
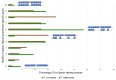
The Fusarium fujikuroi species complex (FFSC) includes socioeconomically important pathogens that cause disease for numerous crops and synthesize a variety of secondary metabolites that can contaminate feedstocks and food. Here, we used comparative genomics to elucidate processes underlying the ability of pine-associated and grass-associated FFSC species to colonize tissues of their respective plant hosts. We characterized the identity, possible functions, evolutionary origins, and chromosomal positions of the host-range-associated genes encoded by the two groups of fungi. The 72 and 47 genes identified as unique to the respective genome groups were potentially involved in diverse processes, ranging from transcription, regulation, and substrate transport through to virulence/pathogenicity. Most genes arose early during the evolution of Fusarium/FFSC and were only subsequently retained in some lineages, while some had origins outside Fusarium. Although differences in the densities of these genes were especially noticeable on the conditionally dispensable chromosome of F. temperatum (representing the grass-associates) and F. circinatum (representing the pine-associates), the host-range-associated genes tended to be located towards the subtelomeric regions of chromosomes. Taken together, these results demonstrate that multiple mechanisms drive the emergence of genes in the grass- and pine-associated FFSC taxa examined. It also highlighted the diversity of the molecular processes potentially underlying niche-specificity in these and other Fusarium species.
Keywords: Fusarium circinatum; Fusarium fujikuroi species complex; Fusarium temperatum; comparative genomics; horizontal gene transfer; host-specificity; pitch canker; subtelomeres.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures

References
-
- Geiser D.M., Aoki T., Bacon C.W., Baker S.E., Bhattacharyya M.K., Brandt M.E., Brown D.W., Burgess L.W., Chulze S., Coleman J.J., et al. One fungus, one name: Defining the genus Fusarium in a scientifically robust way that preserves longstanding use. Phytopathology. 2013;103:400–408. doi: 10.1094/PHYTO-07-12-0150-LE. - DOI - PubMed
-
- Geiser D.M., Al-Hatmi A.M., Aoki T., Arie T., Balmas V., Barnes I., Bergstrom G.C., Bhattacharyya M.K., Blomquist C.L., Bowden R.L. Phylogenomic analysis of a 55.1-kb 19-gene dataset resolves a monophyletic Fusarium that includes the Fusarium solani species complex. Phytopathology. 2021;111:1064–1079. doi: 10.1094/PHYTO-08-20-0330-LE. - DOI - PubMed
-
- Chang D.C., Grant G.B., O’Donnell K., Wannemuehler K.A., Noble-Wang J., Rao C.Y., Jacobson L.M., Crowell C.S., Sneed R.S., Lewil F.M.T., et al. Multistate outbreak of Fusarium keratitis associated with use of a contact lens solution. Am. J. Ophthalmol. 2006;142:896–897. doi: 10.1016/j.ajo.2006.09.010. - DOI - PubMed
-
- Sutton D.A., Brandt M.E. In: Fusarium and Other Opportunistic Hyaline Fungi. 10th ed. Versalovic J., Carroll K.C., Funke G., Jorgenson J.H., Landry M.L., Warnock D.W., editors. ASM Press; Washington, DC, USA: 2011.
-
- O’Donnell K., Sutton D.A., Rinaldi M.G., Magnon K.C., Cox P.A., Revankar S.G., Sanche S., Geiser D.M., Juba J.H., Van Burik J.-A.H. Genetic diversity of human pathogenic members of the Fusarium oxysporum complex inferred from multilocus DNA sequence data and amplified fragment length polymorphism analyses: Evidence for the recent dispersion of a geographically widespread clonal lineage and nosocomial origin. J. Clin. Microbiol. 2004;42:5109–5120. - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

