In Vitro Evaluation and Mitigation of Niclosamide's Liabilities as a COVID-19 Treatment
- PMID: 36016172
- PMCID: PMC9412300
- DOI: 10.3390/vaccines10081284
In Vitro Evaluation and Mitigation of Niclosamide's Liabilities as a COVID-19 Treatment
Abstract
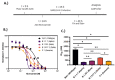
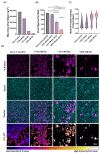
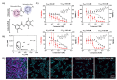
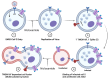
Niclosamide, an FDA-approved oral anthelmintic drug, has broad biological activity including anticancer, antibacterial, and antiviral properties. Niclosamide has also been identified as a potent inhibitor of SARS-CoV-2 infection in vitro, generating interest in its use for the treatment or prevention of COVID-19. Unfortunately, there are several potential issues with using niclosamide for COVID-19, including low bioavailability, significant polypharmacology, high cellular toxicity, and unknown efficacy against emerging SARS-CoV-2 variants of concern. In this study, we used high-content imaging-based immunofluorescence assays in two different cell models to assess these limitations and evaluate the potential for using niclosamide as a COVID-19 antiviral. We show that despite promising preliminary reports, the antiviral efficacy of niclosamide overlaps with its cytotoxicity giving it a poor in vitro selectivity index for anti-SARS-CoV-2 inhibition. We also show that niclosamide has significantly variable potency against the different SARS-CoV-2 variants of concern and is most potent against variants with enhanced cell-to-cell spread including the B.1.1.7 (alpha) variant. Finally, we report the activity of 33 niclosamide analogs, several of which have reduced cytotoxicity and increased potency relative to niclosamide. A preliminary structure-activity relationship analysis reveals dependence on a protonophore for antiviral efficacy, which implicates nonspecific endolysosomal neutralization as a dominant mechanism of action. Further single-cell morphological profiling suggests niclosamide also inhibits viral entry and cell-to-cell spread by syncytia. Altogether, our results suggest that niclosamide is not an ideal candidate for the treatment of COVID-19, but that there is potential for developing improved analogs with higher clinical translational potential in the future.
Keywords: COVID-19; SARS-CoV-2; drug repurposing; niclosamide; polypharmacology.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Update of
-
In Vitro Evaluation and Mitigation of Niclosamide's Liabilities as a COVID-19 Treatment.bioRxiv [Preprint]. 2022 Jul 13:2022.06.24.497526. doi: 10.1101/2022.06.24.497526. bioRxiv. 2022. Update in: Vaccines (Basel). 2022 Aug 09;10(8):1284. doi: 10.3390/vaccines10081284. PMID: 35860224 Free PMC article. Updated. Preprint.
Similar articles
-
In Vitro Evaluation and Mitigation of Niclosamide's Liabilities as a COVID-19 Treatment.bioRxiv [Preprint]. 2022 Jul 13:2022.06.24.497526. doi: 10.1101/2022.06.24.497526. bioRxiv. 2022. Update in: Vaccines (Basel). 2022 Aug 09;10(8):1284. doi: 10.3390/vaccines10081284. PMID: 35860224 Free PMC article. Updated. Preprint.
-
Niclosamide shows strong antiviral activity in a human airway model of SARS-CoV-2 infection and a conserved potency against the Alpha (B.1.1.7), Beta (B.1.351) and Delta variant (B.1.617.2).PLoS One. 2021 Dec 2;16(12):e0260958. doi: 10.1371/journal.pone.0260958. eCollection 2021. PLoS One. 2021. PMID: 34855904 Free PMC article.
-
Identification of oral therapeutics using an AI platform against the virus responsible for COVID-19, SARS-CoV-2.Front Pharmacol. 2023 Dec 22;14:1297924. doi: 10.3389/fphar.2023.1297924. eCollection 2023. Front Pharmacol. 2023. PMID: 38186640 Free PMC article.
-
Broad Spectrum Antiviral Agent Niclosamide and Its Therapeutic Potential.ACS Infect Dis. 2020 May 8;6(5):909-915. doi: 10.1021/acsinfecdis.0c00052. Epub 2020 Mar 10. ACS Infect Dis. 2020. PMID: 32125140 Free PMC article. Review.
-
Niclosamide-A promising treatment for COVID-19.Br J Pharmacol. 2022 Jul;179(13):3250-3267. doi: 10.1111/bph.15843. Epub 2022 Apr 11. Br J Pharmacol. 2022. PMID: 35348204 Free PMC article. Review.
Cited by
-
Salicylanilides and Their Anticancer Properties.Int J Mol Sci. 2023 Jan 15;24(2):1728. doi: 10.3390/ijms24021728. Int J Mol Sci. 2023. PMID: 36675241 Free PMC article. Review.
-
Transforming Niclosamide through Nanotechnology: A Promising Approach for Long COVID Management.Small. 2025 Jul;21(27):e2410345. doi: 10.1002/smll.202410345. Epub 2025 May 19. Small. 2025. PMID: 40384184 Free PMC article. Review.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

