The Role of Lebanon in the COVID-19 Butterfly Effect: The B.1.398 Example
- PMID: 36016262
- PMCID: PMC9412248
- DOI: 10.3390/v14081640
The Role of Lebanon in the COVID-19 Butterfly Effect: The B.1.398 Example
Abstract
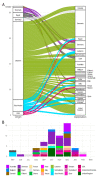
In the present study, we provide a retrospective genomic surveillance of the SARS-CoV-2 pandemic in Lebanon; we newly sequence the viral genomes of 200 nasopharyngeal samples collected between July 2020 and February 2021 from patients in different regions of Lebanon and from travelers crossing the Lebanese-Syrian border, and we also analyze the Lebanese genomic dataset available at GISAID. Our results show that SARS-CoV-2 infections in Lebanon during this period were shaped by the turnovers of four dominant SARS-CoV-2 lineages, with B.1.398 being the first to thoroughly dominate. Lebanon acted as a dispersal center of B.1.398 to other countries, with intercontinental transmissions being more common than within-continent. Within the country, the district of Tripoli, which was the source of 43% of the total B.1.398 sequences in our study, was identified as being an important source of dispersal in the country. In conclusion, our findings exemplify the butterfly effect, by which a lineage that emerges in a small area can be spread around the world, and highlight the potential role of developing countries in the emergence of new variants.
Keywords: B.1.398; Lebanon; SARS-CoV-2; dispersal; lineages.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures




References
-
- Our World in Data. [(accessed on 30 March 2022)]. Available online: https://ourworldindata.org/explorers/coronavirus-data-explorer.
-
- Pango Lineages: Latest Epidemiological Lineages of SARS-CoV-2. [(accessed on 30 March 2022)]. Available online: https://cov-lineages.org/
-
- WHO: Tracking SARS-CoV-2 Variants. [(accessed on 30 March 2022)]. Available online: https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

