Characterization of a Human Sapovirus Genotype GII.3 Strain Generated by a Reverse Genetics System: VP2 Is a Minor Structural Protein of the Virion
- PMID: 36016271
- PMCID: PMC9414370
- DOI: 10.3390/v14081649
Characterization of a Human Sapovirus Genotype GII.3 Strain Generated by a Reverse Genetics System: VP2 Is a Minor Structural Protein of the Virion
Abstract
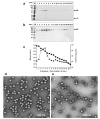
We devised a reverse genetics system to generate an infectious human sapovirus (HuSaV) GII.3 virus. Capped/uncapped full-length RNAs derived from HuSaV GII.3 AK11 strain generated by in vitro transcription were used to transfect HuTu80 human duodenum carcinoma cells; infectious viruses were recovered from the capped RNA-transfected cells and passaged in the cells. Genome-wide analyses indicated no nucleotide sequence change in the virus genomes in the cell-culture supernatants recovered from the transfection or those from the subsequent infection. No virus growth was detected in the uncapped RNA-transfected cells, suggesting that the 5'-cap structure is essential for the virus' generation and replication. Two types of virus particles were purified from the cell-culture supernatant. The complete particles were 39.2-nm-dia., at 1.350 g/cm3 density; the empty particles were 42.2-nm-dia. at 1.286 g/cm3. Two proteins (58-kDa p58 and 17-kDa p17) were detected from the purified particles; their molecular weight were similar to those of VP1 (~60-kDa) and VP2 (~16-kDa) of AK11 strain deduced from their amino acids (aa) sequences. Protein p58 interacted with HuSaV GII.3-VP1-specific antiserum, suggesting that p58 is HuSaV VP1. A total of 94 (57%) aa of p17 were identified by mass spectrometry; the sequences were identical to those of VP2, indicating that the p17 is the VP2 of AK11. Our new method produced infectious HuSaVs and demonstrated that VP2 is the minor protein of the virion, suggested to be involved in the HuSaV assembly.
Keywords: HuSaV; VP1; VP2; genogroup GII.3; human sapovirus; reverse genetics system; virus particle.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

