PEDV: Insights and Advances into Types, Function, Structure, and Receptor Recognition
- PMID: 36016366
- PMCID: PMC9416423
- DOI: 10.3390/v14081744
PEDV: Insights and Advances into Types, Function, Structure, and Receptor Recognition
Abstract
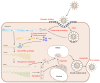
Porcine epidemic diarrhea virus (PEDV) has been endemic in most parts of the world since its emergence in the 1970s. It infects the small intestine and intestinal villous cells, spreads rapidly, and causes infectious intestinal disease characterized by vomiting, diarrhea, and dehydration, leading to high mortality in newborn piglets and causing massive economic losses to the pig industry. The entry of PEDV into cells is mediated by the binding of its spike protein (S protein) to a host cell receptor. Here, we review the structure of PEDV, its strains, and the structure and function of the S protein shared by coronaviruses, and summarize the progress of research on possible host cell receptors since the discovery of PEDV.
Keywords: PEDV receptor; PEDV variant strain; porcine epidemic diarrhea virus (PEDV); spike protein (S protein).
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures



References
-
- Cavanagh D. Nidovirales: A new order comprising Coronaviridae and Arteriviridae. Arch. Virol. 1997;142:629–633. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

