Stabilisation of Viral Membrane Fusion Proteins in Prefusion Conformation by Structure-Based Design for Structure Determination and Vaccine Development
- PMID: 36016438
- PMCID: PMC9415420
- DOI: 10.3390/v14081816
Stabilisation of Viral Membrane Fusion Proteins in Prefusion Conformation by Structure-Based Design for Structure Determination and Vaccine Development
Abstract
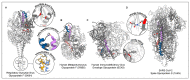
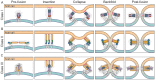
The membrane surface of enveloped viruses contains dedicated proteins enabling the fusion of the viral with the host cell membrane. Working with these proteins is almost always challenging because they are membrane-embedded and naturally metastable. Fortunately, based on a range of different examples, researchers now have several possibilities to tame membrane fusion proteins, making them amenable for structure determination and immunogen generation. This review describes the structural and functional similarities of the different membrane fusion proteins and ways to exploit these features to stabilise them by targeted mutational approaches. The recent determination of two herpesvirus membrane fusion proteins in prefusion conformation holds the potential to apply similar methods to this group of viral fusogens. In addition to a better understanding of the herpesviral fusion mechanism, the structural insights gained will help to find ways to further stabilise these proteins using the methods described to obtain stable immunogens that will form the basis for the development of the next generation of vaccines and antiviral drugs.
Keywords: fusion mechanism; fusogen; glycoprotein B; herpesvirus; membrane fusion; postfusion; prefusion; protein stabilisation; structure-based design; viral fusion protein.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

