Methylation-sensitive high-resolution melting analysis of the USP44 promoter can detect early-stage hepatocellular carcinoma in blood samples
- PMID: 36016503
- PMCID: PMC9712700
- DOI: 10.5483/BMBRep.2022.55.11.110
Methylation-sensitive high-resolution melting analysis of the USP44 promoter can detect early-stage hepatocellular carcinoma in blood samples
Abstract
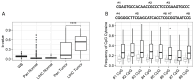
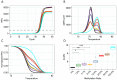
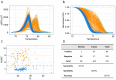
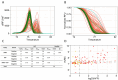
Hepatocellular carcinoma (HCC) is dangerous cancer that often evades early detection because it is asymptomatic and an effective detection method is lacking. For people with chronic liver inflammation who are at high risk of developing HCC, a sensitive detection method for HCC is needed. In a meta-analysis of The Cancer Genome Atlas pan-cancer methylation database, we identified a CpG island in the USP44 promoter that is methylated specifically in HCC. We developed methylation-sensitive high-resolution melting (MS-HRM) analysis to measure the methylation levels of the USP promoter in cell-free DNA isolated from patients. Our MS-HRM assay correctly identified 40% of patients with early-stage HCC, whereas the α-fetoprotein test, which is currently used to detect HCC, correctly identified only 25% of early-stage HCC patients. These results demonstrate that USP44 MS-HRM analysis is suitable for HCC surveillance. [BMB Reports 2022; 55(11): 553-558].
Conflict of interest statement
The authors have no conflicting interests.
Figures
References
-
- Akinyemiju T, Abera S, Ahmed M, et al. The burden of primary liver cancer and underlying etiologies from 1990 to 2015 at the global, regional, and national level: results from the global burden of disease study 2015. JAMA Oncol. 2017;3:1683–1691. doi: 10.1001/jamaoncol.2017.3055. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

