Temporal variations of human and animal Rotavirus A genotypes in surface water used for drinking water production
- PMID: 36016785
- PMCID: PMC9395708
- DOI: 10.3389/fmicb.2022.912147
Temporal variations of human and animal Rotavirus A genotypes in surface water used for drinking water production
Abstract
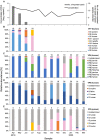
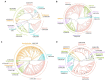
Rotavirus is a major cause of gastroenteritis among infants and children. In this study, nested PCR assays were developed to amplify partial regions of the VP7, VP4, and VP6 genes of Rotavirus A (RVA) for amplicon-based Illumina MiSeq sequencing to investigate RVA genotypes in environmental water samples. Eight sets of inner primers were first designed and screened for use in the nested PCR assays, and four sets of them could produce amplicons. Six sets of outer primers were then designed and combined with the four sets of inner primers that worked. The assays were evaluated for sensitivity using raw water samples collected from one drinking water treatment plant between April 2019 and March 2020 (Sample Set 1; N = 12) and seven DWTPs between 2018 and 2020 (Sample Set 2; N = 18). In total, 43 amplicons from Set 1 were sequenced and diverse sequences from human, porcine, bovine, equine, and feline RVA were observed. Human G8, G3, and G2 genotypes were obtained, with G8 predominant (relative abundance, 36-87%) in samples taken during the rotavirus epidemic season between April and June. Porcine G5, G11, and G4, and bovine G10 and G6 genotypes were also detected. VP4 sequence analysis revealed that the human P[8] genotype was present throughout the year, whereas P[4] and P[9] were present only in the epidemic season. The vaccine strains P[5] and P[8] (RotaTeq®) were also detected. Our approach enables the identification of prevalent human and animal RVA genotypes and their host species that potentially caused fecal contamination in water sources.
Keywords: drinking water source; massive parallel sequencing; molecular epidemiology; rotavirus; surface water.
Copyright © 2022 Miura, Kadoya, Takino, Sano and Akiba.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
Molecular surveillance of rotavirus A associated with diarrheic calves from the Republic of Korea and full genomic characterization of bovine-porcine reassortant G5P[7] strain.Infect Genet Evol. 2022 Jun;100:105266. doi: 10.1016/j.meegid.2022.105266. Epub 2022 Mar 9. Infect Genet Evol. 2022. PMID: 35276340
-
One-step multiplex real-time RT-PCR assay for detecting and genotyping wild-type group A rotavirus strains and vaccine strains (Rotarix® and RotaTeq®) in stool samples.PeerJ. 2016 Jan 11;4:e1560. doi: 10.7717/peerj.1560. eCollection 2016. PeerJ. 2016. PMID: 26839745 Free PMC article.
-
Pepper mild mottle virus intended for use as a process indicator for drinking water treatment: Present forms and quantitative relations to norovirus and rotavirus in surface water.Water Res. 2024 Jun 15;257:121713. doi: 10.1016/j.watres.2024.121713. Epub 2024 May 10. Water Res. 2024. PMID: 38733963
-
Detection and genotyping of human rotavirus VP4 and VP7 genes by reverse transcriptase PCR and reverse hybridization.J Clin Microbiol. 2009 Sep;47(9):2704-12. doi: 10.1128/JCM.00378-09. Epub 2009 Jun 24. J Clin Microbiol. 2009. PMID: 19553575 Free PMC article.
-
Porcine group A rotaviruses with heterogeneous VP7 and VP4 genotype combinations can be found together with enteric bacteria on Belgian swine farms.Vet Microbiol. 2014 Aug 6;172(1-2):23-34. doi: 10.1016/j.vetmic.2014.04.002. Epub 2014 Apr 13. Vet Microbiol. 2014. PMID: 24837191 Review.
Cited by
-
Rotavirus in Water Environments: A Systematic Review and Meta-Analysis.Environ Health Insights. 2024 Oct 18;18:11786302241276667. doi: 10.1177/11786302241276667. eCollection 2024. Environ Health Insights. 2024. PMID: 39439598 Free PMC article. Review.
References
-
- Bisseux M., Debroas D., Mirand A., Archimbaud C., Peigue-Lafeuille H., Bailly J.-L., et al. . (2020). Monitoring of enterovirus diversity in wastewater by ultra-deep sequencing: an effective complementary tool for clinical enterovirus surveillance. Water Res. 169:115246. doi: 10.1016/j.watres.2019.115246, PMID: - DOI - PubMed
-
- Bordería A. V., Isakov O., Moratorio G., Henningsson R., Agüera-González S., Organtini L., et al. . (2015). Group selection and contribution of minority variants during virus adaptation determines virus fitness and phenotype. PLoS Pathog. 11:e1004838. doi: 10.1371/journal.ppat.1004838, PMID: - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

