Spatial organization and early signaling of the B-cell receptor in CLL
- PMID: 36016925
- PMCID: PMC9398492
- DOI: 10.3389/fimmu.2022.953660
Spatial organization and early signaling of the B-cell receptor in CLL
Abstract
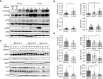
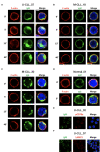
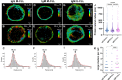
Most chronic lymphocytic leukemia (CLL) clones express B-cell receptors (BcR) of both IgM/IgD isotypes; however, 5%-10% of CLL cases express isotype-switched immunoglobulin G (IgG). The early signaling and spatial patterning of the various BcRs at steady state and after activation are still fully unresolved. Herein, we show higher expression of the BcR signalosome elements and a more robust constitutive cell-intrinsic proximal BcR signaling in CLL with unmutated IGHV expressing IgM isotype (IgM U-CLL), compared with IGHV-mutated CLL (M-CLL) expressing either IgM or IgG isotypes. IgM in U-CLL is frequently located in the membrane plane in polarized patches, occasionally in caps, and sometimes inside the cells. Among M-CLL, IgM is scattered laterally in the membrane plane in a similar pattern as seen in normal B cells, whereas IgG is dispersed around the cell membrane in smaller clusters than in IgM U-CLL. Upon BcR engagement, both IgG and IgM expressing M-CLL showed attenuated signaling and only slight spatial reorganization dynamics of BcR microclusters and internalization, compared with the extensive reorganization and internalization of the BcR in IgM expressing U-CLL. The global gene signature of IgG M-CLL was closely related to that of IgM M-CLL rather than IgM U-CLL. Overall, we report fundamental differences in the basal composition, biochemical status, and spatial organization of the BcR in the three examined immunogenetic CLL subtypes that correlate with their clinical behavior. On the basis of our findings, IgG class-switched M-CLL likely represents the same disease as IgM M-CLL rather than a different biological and/or clinical entity.
Keywords: BCR signaling; CLL (chronic lymphocytic leukemia); IGHV mutational status; IgG; IgM; d-STORM.
Copyright © 2022 Shorer Arbel, Bronstein, Dadosh, Kamdjou, Tsuriel, Shapiro, Katz and Herishanu.
Conflict of interest statement
YH received honoraria from AstraZeneca, Janssen, AbbeVie, and Medison for work unrelated to the present study. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
-
- Herishanu Y, Pérez-Galán P, Liu D, Biancotto A, Pittaluga S, Vire B, et al. . The lymph node microenvironment promotes b-cell receptor signaling, NF-κB activation, and tumor proliferation in chronic lymphocytic leukemia. Blood (2011) 117(2):563–74. doi: 10.1182/blood-2010-05-284984 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

