Identification and Analysis of Long Non-Coding RNAs Related to UV-B-Induced Anthocyanin Biosynthesis During Blood-Fleshed Peach (Prunus persica) Ripening
- PMID: 36017497
- PMCID: PMC9395590
- DOI: 10.3389/fgene.2022.932207
Identification and Analysis of Long Non-Coding RNAs Related to UV-B-Induced Anthocyanin Biosynthesis During Blood-Fleshed Peach (Prunus persica) Ripening
Abstract
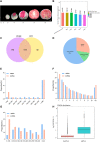
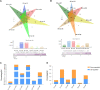
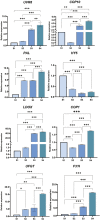
Blood flesh is a key fruit trait in peaches (Prunus persica) and can be attributed to the accumulation of anthocyanins. The roles of long non-coding RNAs (lncRNAs) have been highlighted by multiple studies in regulating fruit ripening, anthocyanin accumulation, and abiotic stress responses in many flowering plants. Such regulatory functions of lncRNAs in Prunus persica, nonetheless, have not been reported. In this research, we sequenced and analyzed the complete transcriptome of C3-20 (a blood-fleshed peach) fruit at four developmental stages. Analyses of the correlated genes and differentially expressed lncRNA target genes helped to forecast lncRNAs' possible functions. The RNA-seq data were generated using high-throughput sequencing. In total, 17,456 putative lncRNAs, including 4,800 intergenic lncRNAs, 2,199 antisense lncRNAs, and 10,439 intronic lncRNAs were discovered, of which 4,871 differentially expressed lncRNAs (DE-lncRNAs) were annotated in the fruit developmental processes. The target genes of these DE-lncRNAs and their regulatory relationship identifying 21,795 cis-regulated and 18,271 trans-regulated targets of the DE-lncRNAs were in a similar way predicted by us. The enriched GO terms for the target genes included anthocyanin biosynthesis. Flavonoid biosynthesis and plant hormone signal transduction were also included in the enriched KEGG pathways. Co-expression network construction demonstrated that the highly expressed genes might co-regulate multiple other genes associated with auxin signal transduction and take effect in equal pathways. We discovered that lncRNAs, including LNC_000987, LNC_000693, LNC_001323, LNC_003610, LNC_001263, and LNC_003380, correlated with fruit that ripened and could take part in ethylene biosynthesis and metabolism and the ABA signaling pathway. Several essential transcription factors, such as ERFs, WRKY70, NAC56, and NAC72, may in a similar way regulate fruit ripening. Three DE-lncRNAs, XLOC_011933, XLOC_001865, and XLOC_042291, are involved in UV-B-induced anthocyanin biosynthesis and positively regulating UVR8 and COP10, were identified and characterized. Our discovery and characterization of XLOC_011933, XLOC_001865, and XLOC_042291 provide a more precise understanding and preliminarily establishes a theoretical framework for UV-B-induced flesh anthocyanin biosynthesis. This phenomenon might encourage more in-depth investigations to study the molecular mechanisms underlying peach flesh coloring.
Keywords: UV-B; anthocyanin biosynthesis; blood-fleshed peach; lncRNAs; ripening.
Copyright © 2022 Zhang, Zhang, Wang, Ye, Liu, Song, Du, Cao, Song, Xiao, Liu, Zhang, Song, Yang, Meng and Wu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



Similar articles
-
Genome-wide identification and characterization of long non-coding RNAs involved in fruit ripening and the climacteric in Cucumis melo.BMC Plant Biol. 2019 Aug 22;19(1):369. doi: 10.1186/s12870-019-1942-4. BMC Plant Biol. 2019. PMID: 31438855 Free PMC article.
-
Gene regulation of anthocyanin biosynthesis in two blood-flesh peach (Prunus persica (L.) Batsch) cultivars during fruit development.J Zhejiang Univ Sci B. 2014 Sep;15(9):809-19. doi: 10.1631/jzus.B1400086. J Zhejiang Univ Sci B. 2014. PMID: 25183035 Free PMC article.
-
Genome-wide identification and characterization of long noncoding RNAs during peach (Prunus persica) fruit development and ripening.Sci Rep. 2022 Jun 30;12(1):11044. doi: 10.1038/s41598-022-15330-3. Sci Rep. 2022. PMID: 35773470 Free PMC article.
-
The ncRNAs Involved in the Regulation of Abiotic Stress-Induced Anthocyanin Biosynthesis in Plants.Antioxidants (Basel). 2023 Dec 28;13(1):55. doi: 10.3390/antiox13010055. Antioxidants (Basel). 2023. PMID: 38247480 Free PMC article. Review.
-
Molecular Control by Non-coding RNAs During Fruit Development: From Gynoecium Patterning to Fruit Ripening.Front Plant Sci. 2018 Nov 30;9:1760. doi: 10.3389/fpls.2018.01760. eCollection 2018. Front Plant Sci. 2018. PMID: 30555499 Free PMC article. Review.
Cited by
-
Integrated Metabolome, Transcriptome and Long Non-Coding RNA Analysis Reveals Potential Molecular Mechanisms of Sweet Cherry Fruit Ripening.Int J Mol Sci. 2024 Sep 12;25(18):9860. doi: 10.3390/ijms25189860. Int J Mol Sci. 2024. PMID: 39337346 Free PMC article.
-
SoNAC72-SoMYB44/SobHLH130 module contributes to flower color fading via regulating anthocyanin biosynthesis by directly binding to the SoUFGT1 promoter in lilac (Syringa oblata).Hortic Res. 2024 Nov 21;12(3):uhae326. doi: 10.1093/hr/uhae326. eCollection 2025 Mar. Hortic Res. 2024. PMID: 40046329 Free PMC article.
-
Chromosome-scale genome, together with transcriptome and metabolome, provides insights into the evolution and anthocyanin biosynthesis of Rubus rosaefolius Sm. (Rosaceae).Hortic Res. 2024 Mar 2;11(4):uhae064. doi: 10.1093/hr/uhae064. eCollection 2024 Apr. Hortic Res. 2024. PMID: 38689697 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Miscellaneous

