Delineating the Spread and Prevalence of SARS-CoV-2 Omicron Sublineages (BA.1-BA.5) and Deltacron Using Wastewater in the Western Cape, South Africa
- PMID: 36017801
- PMCID: PMC9574669
- DOI: 10.1093/infdis/jiac356
Delineating the Spread and Prevalence of SARS-CoV-2 Omicron Sublineages (BA.1-BA.5) and Deltacron Using Wastewater in the Western Cape, South Africa
Abstract
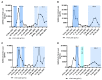
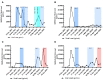
This study was one of the first to detect Omicron sublineages BA.4 and BA.5 in wastewater from South Africa. Spearman rank correlation analysis confirmed a strong positive correlation between severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) viral RNA in wastewater samples and clinical cases (r = 0.7749, P < .0001). SARS-CoV-2 viral load detected in wastewater, resulting from the Delta-driven third wave, was significantly higher than during the Omicron-driven fourth wave. Whole-genome sequencing confirmed presence of Omicron lineage defining mutations in wastewater with the first occurrence reported 23 November 2021 (BA.1 predominant). The variant spread rapidly, with prevalence of Omicron-positive wastewater samples rising to >80% by 10 January 2022 with BA.2 as the predominant sublineage by 10 March 2022, whilst on 18 April 2022 BA.4 and BA.5 were detected in selected wastewater sites. These findings demonstrate the value of wastewater-based epidemiology to monitor the spatiotemporal spread and potential origin of new Omicron sublineages.
Keywords: B.1.5.9 (Omicron); B.1.617.2 (Delta) lineages; BA.1; BA.2; BA.3; SARS-CoV-2; and BA.5; genotyping; wastewater-based epidemiology.
© The Author(s) 2022. Published by Oxford University Press on behalf of Infectious Diseases Society of America.
Conflict of interest statement
Potential conflicts of interest. All authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed. The authors have no conflicts of interest to declare.
Figures




References
-
- World Health Organization . Update on Omicron, 2021. https://www.who.int/news/item/28-11-2021-update-on-omicron. [Accessed: 16 March 2022].
-
- Iuliano AD, Brunkard JM, Boehmer TK, et al. . Trends in disease severity and health care utilization during the early Omicron variant period compared with previous SARS-CoV-2 high transmission periods—United States, December 2020–January 2022. MMWR Morb Mortal Wkly Rep 2022; 71:146–52. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

