GPCR structural characterization by NMR spectroscopy in solution
- PMID: 36017890
- PMCID: PMC9828178
- DOI: 10.3724/abbs.2022106
GPCR structural characterization by NMR spectroscopy in solution
Abstract
In the human proteome, 826 G-protein-coupled receptors (GPCRs) interact with extracellular stimuli to initiate cascades of intracellular signaling. Determining conformational dynamics and intermolecular interactions are key to understand GPCR function as a basis for drug design. X-ray crystallography and cryo-electron microscopy (cryo-EM) contribute molecular architectures of GPCRs and GPCR-signaling complexes. NMR spectroscopy is complementary by providing information on the dynamics of GPCR structures at physiological temperature. In this review, several NMR approaches in use to probe GPCR dynamics and intermolecular interactions are discussed. The topics include uniform stable-isotope labeling, amino acid residue-selective stable-isotope labeling, site-specific labeling by genetic engineering, the introduction of 19F-NMR probes, and the use of paramagnetic nitroxide spin labels. The unique information provided by NMR spectroscopy contributes to our understanding of GPCR biology and thus adds to the foundations for rational drug design.
Keywords: G protein-coupled receptor dynamics; GPCR biology; drug development; fluorine-19 NMR; stable-isotope labeling.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
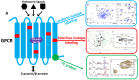
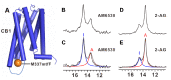
Similar articles
-
GPCR drug discovery: integrating solution NMR data with crystal and cryo-EM structures.Nat Rev Drug Discov. 2019 Jan;18(1):59-82. doi: 10.1038/nrd.2018.180. Epub 2018 Nov 9. Nat Rev Drug Discov. 2019. PMID: 30410121 Free PMC article. Review.
-
G Protein-coupled Receptor (GPCR) Reconstitution and Labeling for Solution Nuclear Magnetic Resonance (NMR) Studies of the Structural Basis of Transmembrane Signaling.Molecules. 2022 Apr 20;27(9):2658. doi: 10.3390/molecules27092658. Molecules. 2022. PMID: 35566006 Free PMC article. Review.
-
NMR analysis of GPCR conformational landscapes and dynamics.Mol Cell Endocrinol. 2019 Mar 15;484:69-77. doi: 10.1016/j.mce.2018.12.019. Epub 2019 Jan 25. Mol Cell Endocrinol. 2019. PMID: 30690069 Review.
-
Design and preparation of the class B G protein-coupled receptors GLP-1R and GCGR for 19 F-NMR studies in solution.FEBS J. 2021 Jul;288(13):4053-4063. doi: 10.1111/febs.15686. Epub 2021 Jan 9. FEBS J. 2021. PMID: 33369025
-
Solution NMR spectroscopy of GPCRs: Residue-specific labeling strategies with a focus on 13C-methyl methionine labeling of the atypical chemokine receptor ACKR3.Methods Cell Biol. 2019;149:259-288. doi: 10.1016/bs.mcb.2018.09.004. Epub 2018 Nov 15. Methods Cell Biol. 2019. PMID: 30616824 Free PMC article.
Cited by
-
Bringing GPCR Structural Biology to Medical Applications: Insights from Both V2 Vasopressin and Mu-Opioid Receptors.Membranes (Basel). 2023 Jun 16;13(6):606. doi: 10.3390/membranes13060606. Membranes (Basel). 2023. PMID: 37367810 Free PMC article. Review.
-
Single-Nucleotide Polymorphisms of TAS2R46 Affect the Receptor Downstream Calcium Regulation in Histamine-Challenged Cells.Cells. 2024 Jul 16;13(14):1204. doi: 10.3390/cells13141204. Cells. 2024. PMID: 39056786 Free PMC article.
References
-
- Pierce KL, Premont RT, Lefkowitz RJ. Seven-transmembrane receptors. Nat Rev Mol Cell Biol. . 2002;3:639–650. doi: 10.1038/nrm908. - DOI - PubMed
-
- Wettschureck N, Offermanns S. Mammalian G proteins and their cell type specific functions. Physiol Rev. . 2005;85:1159–1204. doi: 10.1152/physrev.00003.2005. - DOI - PubMed
-
- Fonin AV, Darling AL, Kuznetsova IM, Turoverov KK, Uversky VN. Multi-functionality of proteins involved in GPCR and G protein signaling: making sense of structure–function continuum with intrinsic disorder-based proteoforms. Cell Mol Life Sci. . 2019;76:4461–4492. doi: 10.1007/s00018-019-03276-1. - DOI - PMC - PubMed
-
- Alexander SP, Kelly E, Marrion N, Peters JA, Benson HE, Faccenda E, Pawson AJ, et al. The concise guide to pharmacology 2015/16: overview. Br J Pharmacol. . 2015;172:5729–5743. doi: 10.1111/bph.13347. - DOI - PMC - PubMed
-
- Sriram K, Insel PA. G protein-coupled receptors as targets for approved drugs: how many targets and how many drugs? Mol Pharmacol. . 2018;93:251–258. doi: 10.1124/mol.117.111062. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

