Characterization of full-length CNBP expanded alleles in myotonic dystrophy type 2 patients by Cas9-mediated enrichment and nanopore sequencing
- PMID: 36018009
- PMCID: PMC9462847
- DOI: 10.7554/eLife.80229
Characterization of full-length CNBP expanded alleles in myotonic dystrophy type 2 patients by Cas9-mediated enrichment and nanopore sequencing
Abstract
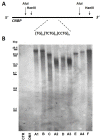
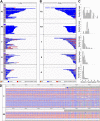
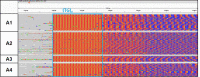
Myotonic dystrophy type 2 (DM2) is caused by CCTG repeat expansions in the CNBP gene, comprising 75 to >11,000 units and featuring extensive mosaicism, making it challenging to sequence fully expanded alleles. To overcome these limitations, we used PCR-free Cas9-mediated nanopore sequencing to characterize CNBP repeat expansions at the single-nucleotide level in nine DM2 patients. The length of normal and expanded alleles can be assessed precisely using this strategy, agreeing with traditional methods, and revealing the degree of mosaicism. We also sequenced an entire ~50 kbp expansion, which has not been achieved previously for DM2 or any other repeat-expansion disorders. Our approach precisely counted the repeats and identified the repeat pattern for both short interrupted and uninterrupted alleles. Interestingly, in the expanded alleles, only two DM2 samples featured the expected pure CCTG repeat pattern, while the other seven presented also TCTG blocks at the 3' end, which have not been reported before in DM2 patients, but confirmed hereby with orthogonal methods. The demonstrated approach simultaneously determines repeat length, structure/motif, and the extent of somatic mosaicism, promising to improve the molecular diagnosis of DM2 and achieve more accurate genotype-phenotype correlations for the better stratification of DM2 patients in clinical trials.
Keywords: CNBP gene; Cas9-mediated enrichment; genetics; genomics; human; myotonic dystrophy type 2; nanopore sequencing; repeat expansion.
© 2022, Alfano, De Antoni et al.
Conflict of interest statement
MA, LD, FC, VV, SM, CD, RM, MD, GN, AB No competing interests declared, MD is a partner of Genartis srl, MR is a partner of Genartis srl, Verona
Figures




References
-
- Bachinski LL, Udd B, Meola G, Sansone V, Bassez G, Eymard B, Thornton CA, Moxley RT, Harper PS, Rogers MT, Jurkat-Rott K, Lehmann-Horn F, Wieser T, Gamez J, Navarro C, Bottani A, Kohler A, Shriver MD, Sallinen R, Wessman M, Zhang S, Wright FA, Krahe R. Confirmation of the type 2 myotonic dystrophy (CCTG)n expansion mutation in patients with proximal myotonic myopathy/proximal myotonic dystrophy of different european origins: A single shared haplotype indicates an ancestral founder effect. American Journal of Human Genetics. 2003;73:835–848. doi: 10.1086/378566. - DOI - PMC - PubMed
-
- Ballester-Lopez A, Koehorst E, Almendrote M, Martínez-Piñeiro A, Lucente G, Linares-Pardo I, Núñez-Manchón J, Guanyabens N, Cano A, Lucia A, Overend G, Cumming SA, Monckton DG, Casadevall T, Isern I, Sánchez-Ojanguren J, Planas A, Rodríguez-Palmero A, Monlleó-Neila L, Pintos-Morell G, Ramos-Fransi A, Coll-Cantí J, Nogales-Gadea G. A DM1 family with interruptions associated with atypical symptoms and late onset but not with A milder phenotype. Human Mutation. 2020;41:420–431. doi: 10.1002/humu.23932. - DOI - PubMed
-
- Botta A, Bonifazi E, Vallo L, Gennarelli M, Garrè C, Salehi L, Iraci R, Sansone V, Meola G, Novelli G. Italian guidelines for molecular analysis in myotonic dystrophies. Acta Myologica. 2006;25:23–33. - PubMed

