Functional diversity of apolipoprotein E: from subcellular localization to mitochondrial function
- PMID: 36018414
- PMCID: PMC9418098
- DOI: 10.1007/s00018-022-04516-7
Functional diversity of apolipoprotein E: from subcellular localization to mitochondrial function
Abstract
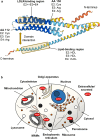
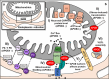
Human apolipoprotein E (APOE), originally known for its role in lipid metabolism, is polymorphic with three major allele forms, namely, APOEε2, APOEε3, and APOEε4, leading to three different human APOE isoforms. The ε4 allele is a genetic risk factor for Alzheimer's disease (AD); therefore, the vast majority of APOE research focuses on its role in AD pathology. However, there is increasing evidence for other functions of APOE through the involvement in other biological processes such as transcriptional regulation, mitochondrial metabolism, immune response, and responsiveness to dietary factors. Therefore, the aim of this review is to provide an overview of the potential novel functions of APOE and their characterization. The detection of APOE in various cell organelles points to previously unrecognized roles in mitochondria and others, although it is actually considered a secretory protein. Furthermore, numerous interactions of APOE with other proteins have been detected, providing indications for new metabolic pathways involving APOE. The present review summarizes the current evidence on APOE beyond its original role in lipid metabolism, to change the perspective and encourage novel approaches to future research on APOE and its isoform-dependent role in the cellular metabolism.
Keywords: Interactome; Mitochondria-associated ER membranes; Protein–protein interactions; Proteolytic fragmentation.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that there is no conflict of interest.
Figures
References
-
- Strittmatter WJ, Saunders AM, Schmechel D, Pericak-Vance M, Enghild J, Salvesen GS, et al. Apolipoprotein E: high-avidity binding to beta-amyloid and increased frequency of type 4 allele in late-onset familial Alzheimer disease. Proc Natl Acad Sci U S A. 1993;90(5):1977–1981. doi: 10.1073/pnas.90.5.1977. - DOI - PMC - PubMed
-
- Martínez-Morillo E, Hansson O, Atagi Y, Bu G, Minthon L, Diamandis EP, et al. Total apolipoprotein E levels and specific isoform composition in cerebrospinal fluid and plasma from Alzheimer’s disease patients and controls. Acta Neuropathol. 2014;127(5):633–643. doi: 10.1007/s00401-014-1266-2. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

