Attribute Analytics Performance Metrics from the MAM Consortium Interlaboratory Study
- PMID: 36018776
- PMCID: PMC9460773
- DOI: 10.1021/jasms.2c00129
Attribute Analytics Performance Metrics from the MAM Consortium Interlaboratory Study
Abstract
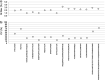
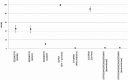
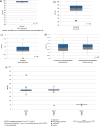
The multi-attribute method (MAM) was conceived as a single assay to potentially replace multiple single-attribute assays that have long been used in process development and quality control (QC) for protein therapeutics. MAM is rooted in traditional peptide mapping methods; it leverages mass spectrometry (MS) detection for confident identification and quantitation of many types of protein attributes that may be targeted for monitoring. While MAM has been widely explored across the industry, it has yet to gain a strong foothold within QC laboratories as a replacement method for established orthogonal platforms. Members of the MAM consortium recently undertook an interlaboratory study to evaluate the industry-wide status of MAM. Here we present the results of this study as they pertain to the targeted attribute analytics component of MAM, including investigation into the sources of variability between laboratories and comparison of MAM data to orthogonal methods. These results are made available with an eye toward aiding the community in further optimizing the method to enable its more frequent use in the QC environment.
Keywords: MAM Consortium; NISTmAb; attribute analytics; multi-attribute method; targeted analytics.
Conflict of interest statement
The authors declare no competing financial interest.
Figures





References
-
- Rogers R; Livingston B; Kerr J; Deng S; Nightlinger N; Scott B; Brady L; Affholter B; Bailey B; Li W; Benchaar S; Luo Q; Peterman S; Prakash A; Wang H; Wheeler K; Bennett P; Balland A.. MS in QC: A Single Multi-attribute Method for Quality Control and Release Testing of Biologics. 10th Symposium on the Practical Applications of Mass Spectrometry in the Biotechnology Industry; Boston (MA), Sep 24–26, 2013.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

