Transcriptome analysis of flower bud identified genes associated with pistil abortions between long branches and spur twigs in apricots (Prunus armeniaca L.)
- PMID: 36018857
- PMCID: PMC9417009
- DOI: 10.1371/journal.pone.0273109
Transcriptome analysis of flower bud identified genes associated with pistil abortions between long branches and spur twigs in apricots (Prunus armeniaca L.)
Abstract
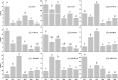
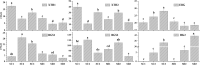
Pistil abortions of flower buds occur frequently in many apricot cultivars, especially in long branches. However, the molecular mechanism underlying pistil abortion in apricots remains unclear. To better understand the molecular mechanism of pistil abortions between long branches and spur twigs, paraffin sections and high-throughput sequencing technology were employed to analyze the expression patterns of genes associated with pistil abortions during later flower bud development stage in 'Shajinhong' apricot. The result of stage III (separation of bud scales) was the critical stage of pistil abortion in apricots. A total of 163 differentially expressed genes were identified as candidate genes related to pistil abortion in long branches. These genes are implicated in programmed cell death, hormone signaling, cell wall degeneration, and the carbohydrate metabolism pathway. The results showed that the up-regulation of gene expression of Xyloglucan endotransglucosylase/hydrolase and β-glucosidase in flower buds might be the direct cause of cell wall breakdown and pistil necrosis in long branches. We hypothesize that there is a molecular relationship between pistil abortion before blooming and cellulose degradation, and then carbohydrate transport in the case of carbon deficiency in long branches. Our work provides new insights into cellulose degradation in abortion pistils and valuable information on flower development in apricots, and also provides a useful reference for cultivation regulation in apricot or other fruit crops.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures









Similar articles
-
Identification of differentially-expressed genes associated with pistil abortion in Japanese apricot by genome-wide transcriptional analysis.PLoS One. 2012;7(10):e47810. doi: 10.1371/journal.pone.0047810. Epub 2012 Oct 16. PLoS One. 2012. PMID: 23091648 Free PMC article.
-
Analyzing Differentially Expressed Genes and Pathways Associated with Pistil Abortion in Japanese Apricot via RNA-Seq.Genes (Basel). 2020 Sep 15;11(9):1079. doi: 10.3390/genes11091079. Genes (Basel). 2020. PMID: 32942711 Free PMC article.
-
Frost tolerance in apricot (Prunus armeniaca L.) receptacle and pistil organs: how is the relationship among amino acids, minerals, and cell death points?Int J Biometeorol. 2021 Dec;65(12):2157-2170. doi: 10.1007/s00484-021-02178-x. Epub 2021 Jul 29. Int J Biometeorol. 2021. PMID: 34324064
-
High-throughput sequencing of small RNAs and analysis of differentially expressed microRNAs associated with pistil development in Japanese apricot.BMC Genomics. 2012 Aug 3;13:371. doi: 10.1186/1471-2164-13-371. BMC Genomics. 2012. PMID: 22863067 Free PMC article.
-
The Multi-Pistil Phenomenon in Higher Plants.Plants (Basel). 2025 Apr 4;14(7):1125. doi: 10.3390/plants14071125. Plants (Basel). 2025. PMID: 40219193 Free PMC article. Review.
References
-
- Cao K, Wang L, Zhu G, Fang W, Chen C. Genetic linkage maps constructing and markers linked with pistil development and double petal gene in peach. Acta Hortic Sin. 2009; 36(2): 179–186. doi: 10.16420/j.issn.0513-353x.2009.02.004 - DOI
-
- Julian C, Herrero M, Rodrigo J. Flower bud differentiation and development in fruiting and non-fruiting shoots in relation to fruit set in apricot (Prunus armeniaca L.). Tree. 2010; 24(5): 833–841. doi: 10.1007/s00468-010-0453-6 - DOI
-
- Rodrigo J, Hormaza JI, Herrero M. Ovary starch reserves and flower development in apricot (Prunus armeniaca). Physiol Plantarum. 2000; 108 (1): 35–41. doi: 10.1034/j.1399-3054.2000.108001035.x - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

