The comparative genomic analysis provides insights into the phylogeny and virulence of tick-borne encephalitis virus vaccine strain Senzhang
- PMID: 36018897
- PMCID: PMC9417034
- DOI: 10.1371/journal.pone.0273565
The comparative genomic analysis provides insights into the phylogeny and virulence of tick-borne encephalitis virus vaccine strain Senzhang
Abstract
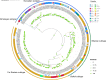
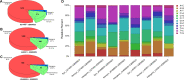
Tick-borne encephalitis virus (TBEV) is one of the most dangerous tick-borne viral pathogens for humans. It can cause severe tick-borne encephalitis (TBE), multiple neurological complications, and death. The European subtype (TBEV-Eu), Siberian subtype (TBEV-Sib), and Far-Eastern subtype (TBEV-FE) are three main TBEV subtypes, causing varying clinical manifestations. Though TBEV-FE is the most virulent TBEV subtype, the degree of variation in the amino acid sequence of TBEV polyprotein is not high, leaving an issue without proper explanation. We performed phylogenic analysis on 243 TBEV strains and then took Senzhang strain as a query strain and representative strains of three major TBEV subtypes as reference strains to perform the comparative genomic analysis, including synteny analysis, SNP analysis, InDel analysis, and multiple sequence alignment of their envelope (E) proteins. The results demonstrated that insertions or deletions of large fragments occurred at the 3' end but not at the 5' end or in the CDS region of TBEV Senzhang strain. In addition, SNP sites are mainly located in the CDS region, with few SNP sites in the non-coding region. Our data highlighted the insertions or deletions of large fragments at the 3' end and SNP sites in the CDS region as genomic properties of the TBEV Senzhang strain compared to representative strains with the main subtypes. These features are probably related to the virulence of the TBEV Senzhang strain and could be considered in future vaccine development and drug target screening for TBEV.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



Similar articles
-
Tick-borne encephalitis in Europe and Russia: Review of pathogenesis, clinical features, therapy, and vaccines.Antiviral Res. 2019 Apr;164:23-51. doi: 10.1016/j.antiviral.2019.01.014. Epub 2019 Jan 31. Antiviral Res. 2019. PMID: 30710567 Review.
-
Tick-borne encephalitis virus in arthropod vectors in the Far East of Russia.Ticks Tick Borne Dis. 2018 May;9(4):824-833. doi: 10.1016/j.ttbdis.2018.01.020. Epub 2018 Feb 8. Ticks Tick Borne Dis. 2018. PMID: 29555424
-
[Phylogenetic analysis and distribution of far eastern tick-borne encephalitis virus subtype (Flaviridae, Flavirus, TBEV-FE) from Asia.].Vopr Virusol. 2019;64(5):250-256. doi: 10.36233/0507-4088-2019-64-5-250-256. Vopr Virusol. 2019. PMID: 32167691 Russian.
-
New genetic lineage within the Siberian subtype of tick-borne encephalitis virus found in Western Siberia, Russia.Infect Genet Evol. 2017 Dec;56:36-43. doi: 10.1016/j.meegid.2017.10.020. Epub 2017 Oct 22. Infect Genet Evol. 2017. PMID: 29069610
-
[Molecular genetic characteristics of tick-borne encephalitis virus].Vopr Virusol. 2007 Sep-Oct;52(5):10-6. Vopr Virusol. 2007. PMID: 18041218 Review. Russian.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

