PRC2-mediated repression is essential to maintain identity and function of differentiated dopaminergic and serotonergic neurons
- PMID: 36026451
- PMCID: PMC9417181
- DOI: 10.1126/sciadv.abo1543
PRC2-mediated repression is essential to maintain identity and function of differentiated dopaminergic and serotonergic neurons
Abstract
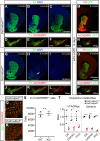
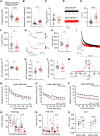
How neurons can maintain cellular identity over an entire life span remains largely unknown. Here, we show that maintenance of identity in differentiated dopaminergic and serotonergic neurons is critically reliant on the Polycomb repressive complex 2 (PRC2). Deletion of the obligate PRC2 component, Eed, in these neurons resulted in global loss of H3K27me3, followed by a gradual activation of genes harboring both H3K27me3 and H3K9me3 modifications. Notably, H3K9me3 was lost at these PRC2 targets before gene activation. Neuronal survival was not compromised; instead, there was a reduction in subtype-specific gene expression and a progressive impairment of dopaminergic and serotonergic neuronal function, leading to behavioral deficits characteristic of Parkinson's disease and anxiety. Single-cell analysis revealed subtype-specific vulnerability to loss of PRC2 repression in dopamine neurons of the substantia nigra. Our study reveals that a PRC2-dependent nonpermissive chromatin state is essential to maintain the subtype identity and function of dopaminergic and serotonergic neurons.
Figures






References
-
- Holmberg J., Perlmann T., Maintaining differentiated cellular identity. Nat. Rev. Genet. 13, 429–439 (2012). - PubMed
-
- Bantignies F., Cavalli G., Cellular memory and dynamic regulation of polycomb group proteins. Curr. Opin. Cell Biol. 18, 275–283 (2006). - PubMed
-
- Di Croce L., Helin K., Transcriptional regulation by Polycomb group proteins. Nat. Struct. Mol. Biol. 20, 1147–1155 (2013). - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

