The protein arginine methyltransferase PRMT9 attenuates MAVS activation through arginine methylation
- PMID: 36028484
- PMCID: PMC9418238
- DOI: 10.1038/s41467-022-32628-y
The protein arginine methyltransferase PRMT9 attenuates MAVS activation through arginine methylation
Erratum in
-
Author Correction: The protein arginine methyltransferase PRMT9 attenuates MAVS activation through arginine methylation.Nat Commun. 2024 Jan 4;15(1):282. doi: 10.1038/s41467-023-44545-9. Nat Commun. 2024. PMID: 38177165 Free PMC article. No abstract available.
Abstract
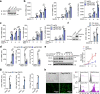
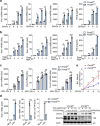
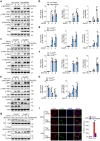
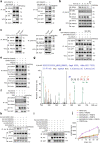
The signaling adaptor MAVS forms prion-like aggregates to activate the innate antiviral immune response after viral infection. However, spontaneous aggregation of MAVS can lead to autoimmune diseases. The molecular mechanism that prevents MAVS from spontaneous aggregation in resting cells has been enigmatic. Here we report that protein arginine methyltransferase 9 targets MAVS directly and catalyzes the arginine methylation of MAVS at the Arg41 and Arg43. In the resting state, this modification inhibits MAVS aggregation and autoactivation of MAVS. Upon virus infection, PRMT9 dissociates from the mitochondria, leading to the aggregation and activation of MAVS. Our study implicates a form of post-translational modification on MAVS, which can keep MAVS inactive in physiological conditions to maintain innate immune homeostasis.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

