PERM1 regulates genes involved in fatty acid metabolism in the heart by interacting with PPARα and PGC-1α
- PMID: 36028747
- PMCID: PMC9418182
- DOI: 10.1038/s41598-022-18885-3
PERM1 regulates genes involved in fatty acid metabolism in the heart by interacting with PPARα and PGC-1α
Abstract
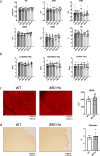
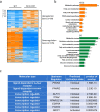
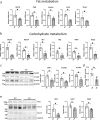
PERM1 (PGC-1/ERR-induced regulator in muscle 1) is a muscle-specific protein induced by PGC-1 and ERRs. Previous studies have shown that PERM1 promotes mitochondrial biogenesis and metabolism in cardiomyocytes in vitro. However, the role of endogenous PERM1 in the heart remains to be investigated with loss-of-function studies in vivo. We report the generation and characterization of systemic Perm1 knockout (KO) mice. The baseline cardiac phenotype of the homozygous Perm1 KO mice appeared normal. However, RNA-sequencing and unbiased pathway analyses showed that homozygous downregulation of PERM1 leads to downregulation of genes involved in fatty acid and carbohydrate metabolism in the heart. Transcription factor binding site analyses suggested that PPARα and PGC-1α are involved in changes in the gene expression profile. Chromatin immunoprecipitation assays showed that PERM1 interacts with the proximal regions of PPAR response elements (PPREs) in endogenous promoters of genes involved in fatty acid oxidation. Co-immunoprecipitation and reporter gene assays showed that PERM1 promoted transcription via the PPRE, partly in a PPARα and PGC-1α dependent manner. These results suggest that endogenous PERM1 is involved in the transcription of genes involved in fatty acid oxidation through physical interaction with PPARα and PGC-1α in the heart in vivo.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Cho Y, Hazen BC, Russell AP, Kralli A. Peroxisome proliferator-activated receptor gamma coactivator 1 (PGC-1)- and estrogen-related receptor (ERR)-induced regulator in muscle 1 (Perm1) is a tissue-specific regulator of oxidative capacity in skeletal muscle cells. J. Biol. Chem. 2013;288:25207–25218. doi: 10.1074/jbc.M113.489674. - DOI - PMC - PubMed
-
- Huss JM, Kopp RP, Kelly DP. Peroxisome proliferator-activated receptor coactivator-1alpha (PGC-1alpha) coactivates the cardiac-enriched nuclear receptors estrogen-related receptor-alpha and -gamma. Identification of novel leucine-rich interaction motif within PGC-1alpha. J. Biol. Chem. 2002;277:40265–40274. doi: 10.1074/jbc.M206324200. - DOI - PubMed
-
- Huss JM, Torra IP, Staels B, Giguere V, Kelly DP. Estrogen-related receptor alpha directs peroxisome proliferator-activated receptor alpha signaling in the transcriptional control of energy metabolism in cardiac and skeletal muscle. Mol. Cell Biol. 2004;24:9079–9091. doi: 10.1128/MCB.24.20.9079-9091.2004. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

