The mIAA7 degron improves auxin-mediated degradation in Caenorhabditiselegans
- PMID: 36029236
- PMCID: PMC9526053
- DOI: 10.1093/g3journal/jkac222
The mIAA7 degron improves auxin-mediated degradation in Caenorhabditiselegans
Abstract
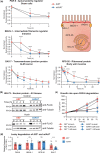
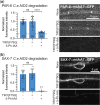
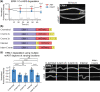
Auxin-inducible degradation is a powerful tool for the targeted degradation of proteins with spatiotemporal control. One limitation of the auxin-inducible degradation system is that not all proteins are degraded efficiently. Here, we demonstrate that an alternative degron sequence, termed mIAA7, improves the efficiency of degradation in Caenorhabditiselegans, as previously reported in human cells. We tested the depletion of a series of proteins with various subcellular localizations in different tissue types and found that the use of the mIAA7 degron resulted in faster depletion kinetics for 5 out of 6 proteins tested. The exception was the nuclear protein HIS-72, which was depleted with similar efficiency as with the conventional AID* degron sequence. The mIAA7 degron also increased the leaky degradation for 2 of the tested proteins. To overcome this problem, we combined the mIAA7 degron with the C. elegans AID2 system, which resulted in complete protein depletion without detectable leaky degradation. Finally, we show that the degradation of ERM-1, a highly stable protein that is challenging to deplete, could be improved further by using multiple mIAA7 degrons. Taken together, the mIAA7 degron further increases the power and applicability of the auxin-inducible degradation system. To facilitate the generation of mIAA7-tagged proteins using CRISPR/Cas9 genome engineering, we generated a toolkit of plasmids for the generation of dsDNA repair templates by PCR.
Keywords: C aenorhabditis elegans; AID system; TIR1; mIAA7; protein degradation.
© The Author(s) 2022. Published by Oxford University Press on behalf of Genetics Society of America.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
