Source of human milk (mother or donor) is more important than fortifier type (human or bovine) in shaping the preterm infant microbiome
- PMID: 36029771
- PMCID: PMC9512671
- DOI: 10.1016/j.xcrm.2022.100712
Source of human milk (mother or donor) is more important than fortifier type (human or bovine) in shaping the preterm infant microbiome
Abstract
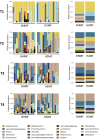
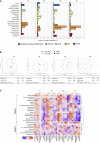
Milk fortifiers help meet the nutritional needs of preterm infants receiving their mother's own milk (MOM) or donor human milk. We conducted a randomized clinical trial (NCT03214822) in 30 very low birth weight premature neonates comparing bovine-derived human milk fortifier (BHMF) versus human-derived fortifier (H2MF). We found that fortifier type does not affect the overall microbiome, although H2MF infants were less often colonized by an unclassified member of Clostridiales Family XI. Secondary analyses show that MOM intake is strongly associated with weight gain and microbiota composition, including Bifidobacterium, Veillonella, and Propionibacterium enrichment. Finally, we show that while oxidative stress (urinary F2-isoprostanes) is not affected by fortifier type or MOM intake, fecal calprotectin is higher in H2MF infants and lower in those consuming more MOM. Overall, the source of human milk (mother versus donor) appears more important than the type of milk fortifier (human versus bovine) in shaping preterm infant gut microbiota.
Keywords: calprotectin; gut inflammation; gut microbiome; human donor milk; human milk fortifiers; mother’s own milk; oxidative stress; very low birth weight infants.
Crown Copyright © 2022. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests M.B.A. has consulted for DSM Nutritional Products and serves on the Malaika Vx and Tiny Health scientific advisory boards. She has received honoraria for speaking at symposia sponsored by Medela, Prolacta Biosciences, and the Institute for Advancement of Breastfeeding and Lactation Education and has contributed without remuneration to online courses on breast milk and the infant microbiome produced by Microbiome Courses. S.V.K. is currently employed by Digbi Health (3T and AI Pvt. Ltd., India), a position taken up after concluding the research presented in this study. These entities had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the article; or decision to submit the article for publication.
Figures



Comment in
-
Reply to SV Kumbhare et al.J Nutr. 2024 May;154(5):1723. doi: 10.1016/j.tjnut.2023.11.029. Epub 2023 Nov 30. J Nutr. 2024. PMID: 38042349 No abstract available.
References
-
- Arslanoglu S., Boquien C.-Y., King C., Lamireau D., Tonetto P., Barnett D., Bertino E., Gaya A., Gebauer C., Grovslien A., et al. Fortification of human milk for preterm infants: update and recommendations of the European milk bank association (EMBA) working group on human milk fortification. Front. Pediatr. 2019;7:76. doi: 10.3389/fped.2019.00076. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

