Biofilms: Formation, Research Models, Potential Targets, and Methods for Prevention and Treatment
- PMID: 36031384
- PMCID: PMC9561771
- DOI: 10.1002/advs.202203291
Biofilms: Formation, Research Models, Potential Targets, and Methods for Prevention and Treatment
Abstract
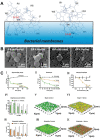
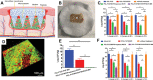
Due to the continuous rise in biofilm-related infections, biofilms seriously threaten human health. The formation of biofilms makes conventional antibiotics ineffective and dampens immune clearance. Therefore, it is important to understand the mechanisms of biofilm formation and develop novel strategies to treat biofilms more effectively. This review article begins with an introduction to biofilm formation in various clinical scenarios and their corresponding therapy. Established biofilm models used in research are then summarized. The potential targets which may assist in the development of new strategies for combating biofilms are further discussed. The novel technologies developed recently for the prevention and treatment of biofilms including antimicrobial surface coatings, physical removal of biofilms, development of new antimicrobial molecules, and delivery of antimicrobial agents are subsequently presented. Finally, directions for future studies are pointed out.
Keywords: biofilms; formation; management; models; targets.
© 2022 The Authors. Advanced Science published by Wiley-VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
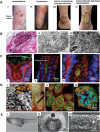


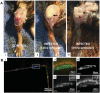

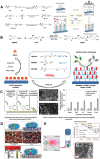


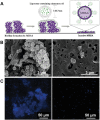
References
-
- Dufour D., Leung V., Lévesque C. M., Endod. Topics 2010, 22, 2.
-
- Costerton J. W., Stewart P. S., Greenberg E. P., Science 1999, 284, 1318. - PubMed
-
- Flemming H.‐C., Wingender J., Nat. Rev. Microbiol. 2010, 8, 623. - PubMed
-
- Kleinman H. K., Philp D., Hoffman M. P., Curr. Opin. Pharmacol. 2003, 14, 526. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
